The KIR Gene Cluster
Authors
Mary Carrington1 and Paul Norman2.Affiliations
Excerpt
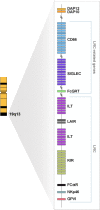
Killer cell immunoglobulin-like receptors (KIRs) are members of a group of regulatory molecules found on subsets of lymphoid cells. They were first identified by their ability to impart some specificity on natural killer (NK) cytolysis. The KIR locus, containing a family of polymorphic and highly homologous genes, maps to chromosome 19q13.4 within the 1 Mb leukocyte receptor complex (LRC). The LRC also encodes the leukocyte Ig-like receptor family (LILR), the leukocyte-associated inhibitory receptor (LAIR) family, and the Fcα receptor. KIR genes are tandemly arrayed over about 150 kb, with the remarkable feature that gene content varies between haplotypes. The discovery of KIR has also imparted an additional function on the human leukocyte antigen (HLA) class I molecules, which are encoded by genes within the major histocompatibility complex (MHC; chromosome 6). Through their interaction with KIR isotypes that inhibit natural killer (NK) cell activity, certain HLA class I molecules are now known to protect healthy cells from spontaneous destruction by NK-cell-mediated cytolysis. Other KIR isotypes stimulate the activity of NK cells. Thus, KIR are likely to play a significant role in the control of the immune response, which would explain the associations observed between certain KIR genes in rheumatoid arthritis, psoriatic arthritis and control of HIV disease progression. The degree of HLA/KIR compatibility may also determine the success rate of haematopoietic cell replacement therapy for certain leukemias.
Introduction
Killer cell immunoglobulin-like receptors (KIRs) are members of a group of regulatory molecules found on subsets of lymphoid cells. They were first identified by their ability to impart some specificity on natural killer (NK) cytolysis (1, 2). The KIR locus, containing a family of polymorphic and highly homologous genes, maps to chromosome 19q13.4 within the 1 Mb leukocyte receptor complex (LRC; Figure 1). The LRC also encodes the leukocyte Ig-like receptor family (LILR; see Box 1 and discussion below), the leukocyte-associated inhibitory receptor (LAIR) family, and the Fcα receptor. KIR genes are tandemly arrayed over about 150 kb, with the remarkable feature that gene content varies between haplotypes (3). The discovery of KIR has also imparted an additional function on the human leukocyte antigen (HLA) class I molecules, which are encoded by genes within the major histocompatibility complex (MHC; chromosome 6). Through their interaction with KIR isotypes that inhibit natural killer (NK) cell activity, certain HLA class I molecules are now known to protect healthy cells from spontaneous destruction by NK-cell-mediated cytolysis. Other KIR isotypes stimulate the activity of NK cells. Thus, KIR are likely to play a significant role in the control of the immune response, which would explain the associations observed between certain KIR genes in rheumatoid arthritis (4), psoriatic arthritis (5) and control of HIV disease progression (6). The degree of HLA/KIR compatibility may also determine the success rate of haematopoietic cell replacement therapy for certain leukemias (7, 8).
Box 1
Unique LILR sequences.
NK cells are an important component of the innate immune system; they participate in early responses against infected or transformed cells by production of cytokines and direct cytotoxicity (9–13). HLA molecules precipitate adaptive aspects of anti-pathogen defense by presenting peptide fragments to immune effector cells (14, 15). Cytotoxic T lymphocytes (CTL) interact with the HLA class I-peptide complex on target cells via the T cell receptor (TCR), which instigates cytolytic activity if the peptide is considered foreign. HLA class I expression can be down-regulated in virally-infected or transformed cells, rendering the cells resistant to cytolysis by CTLs. However, aberrant levels of class I expression can result in spontaneous destruction by NK cells (16–18), a concept originally termed the "missing-self hypothesis" (19). Although neither the ligand nor the direct mode of action of many NK receptors is known, it is widely accepted that normal cells are protected from spontaneous killing when they express an appropriate ligand for an inhibitory receptor expressed by the cytotoxic cell (NK or CTL). NK cells need to discriminate between healthy and infected or transformed cells, corresponding with the observed phenotypic dominance of KIR-mediated inhibition over activation (20–23). Furthermore, inhibition by non-HLA specific NK receptors can override potential activation signals (24).
Studies performed over the last few years have revealed extensive diversity at the KIR gene locus, which stems from both its polygenic and multi-allelic polymorphism (3). As a consequence, there is only a small probability that two randomly selected individuals will have the same KIR genotype (25). KIR gene expression patterns can vary clonally (26), adding yet another layer of complexity to the system. Diversity at the locus may be the result of selection pressures, in a manner analogous to that proposed for the HLA loci. Thus, disease resistance conferred by the KIR locus is likely to vary in a haplotypic manner depending on disease type.
LILR are genetically, structurally and functionally related to KIR and may be their ancestral predecessors (27) (Box 1). Several investigators identified the molecules independently, which has led to inconsistencies in nomenclature systems. The LILRs are also known as ILT [immunoglobulin-like transcript; (28)], LIR [leukocyte inhibitory receptor; (29)], MIR [macrophage inhibitory receptor; (30)] and HM transcripts (31). LILR is the more recently derived and HUGO-endorsed nomenclature. The molecules were identified in binding studies of immune evasion by CMV (29), and in expression screening for human homologues of mouse leukocyte inhibitory receptors (28, 30, 31). Like KIRs, LILRs can interact with HLA class I (29, 32–34), are expressed by a range of immunologically active cells, including NK (31, 35), and have the potential to regulate the immune response through inhibition or activation of cytolytic activity (28, 35–37). LILR genes have been found in a wide variety of species (38–40) and the number of loci appears to be relatively stable (41). The duplicated sub-cluster organization of the LILR gene complex is conserved between humans and chimpanzees (38), unlike KIR haplotypes, which have distinct arrangements that are specific to each of these species (42). LILRs have either two or four extracellular Ig domains and a long or short cytoplasmic tail. Long cytoplasmic tails contain up to four immunoreceptor tyrosine-based inhibitory molecules (ITIM) (43) and therefore have the capacity to inhibit cellular activity. LILR with short cytoplasmic domains can associate with molecules containing ITAMs and contribute to cell activation. LILRA3 (ILT6) is unique in that it does not possess a cytoplasmic domain, and may be secreted (31, 44). Also, much of the LILRA3 gene is missing on some haplotypes due to a 7kb deletion in the region (45). Ligands are not known for all of the LILR, but some, such as LILRB1 (ILT2), can bind to HLA class I (Table 1). Thus, their functions and ligand specificities described so far suggests that LILR and KIR have overlapping and potentially complementary functions.
Table 1
Known LILR, cellular distribution, and ligands.
The LILR region in both human and chimpanzees consists of two sub-clusters of six or seven loci in opposite transcriptional orientation separated by two LAIR genes (27, 38, 46), which encode molecules that interact with epithelial cell adhesion molecule (EpCAM) (47). LILRB3 is highly polymorphic, as 18 variants were observed among only 50 individuals (45, 48). Twelve alleles have been identified for LILRA3 (Table 1) (49).
KIR Nomenclature
About 14 expressed KIR genes have been identified (Table 2) and two systems have been generated for naming them. The most commonly used nomenclature system accounts for their protein structure and consists of four major subdivisions based on two features: the number of extracellular Ig domains (2D or 3D) and characteristics of the cytoplasmic tail (Figure 2) (see PROW and/or the HUGO-endorsed nomenclatures). They have also been named according to the CD nomenclature system as CD158a, CD158b, etc., based on an approximate centromeric–telomeric order of the genes on chromosome 19 (50). Unfortunately, the CD nomenclature does not reflect structure, function, expression or localization (51). This system also presents the possibility of confusion with the monoclonal antibodies, CD158a and CD158b, since each binds to several different KIR molecules. The CD nomenclature is not used routinely, and the Human Genome Organization (HUGO) nomenclature system will be used throughout this report.
Table 2
KIR gene names.
Irrespective of the number of Ig subunits, the cytoplasmic domain of KIR are either long (designated "L") or short ("S"). KIR with long cytoplasmic domains are inhibitory by virtue of the immunoreceptor tyrosine-based inhibition motifs (ITIMs) present in their cytoplasmic domains. Short-tailed KIR transmit activating signals through their interaction with the adaptor molecule, DAP-12 (DNAX activation protein of 12kD; this molecule is also known as killer cell activating receptor-associated protein or KARAP), which contains immunoreceptor tyrosine-based activation motifs (ITAMs) (52, 53). DAP-12 is also a member of the immunoglobulin superfamily and is encoded at the centromeric end of the LRC. ITIMs and ITAMs are characteristic of several immunologically important receptors, such as CD5, CD22 and FcγRII (54).
Two KIR pseudogenes (2DP or 3DP) have been identified (Table 2): 2DP1, which shares high sequence similarity with two-domain KIR genes, and 3DP1, which is similar to 3DL3 in portions of the gene, but may represent an ancestral KIR gene. The pseudogenes have been given various names in the literature, and an alias key and Accession numbers are provided at (www.gene.ucl.ac.uk/nomenclature/genefamily/kir.html).
Allelic variation has been observed for most of the KIR genes and names for alleles at several of the most polymorphic loci have been specified based on nomenclature used for HLA loci (25, 55). The nomenclature for allelic variants is not complete, but a report that goes some way toward addressing this issue is now available (215).
KIR Gene Sequences
Well over 100 KIR sequences have been deposited into either the EMBL or GenBank nucleotide sequence databases. A number of the entries are partial cDNA or genomic sequences and some are identical to a portion of the full-length version. We have generated an alignment of the full-length cDNA sequences (Box 2). Accession numbers of these sequences are provided in Table 2. Each unassigned KIR sequence identified in GenBank was designated as an allele of a specific KIR gene if it differed by <2% from the consensus sequence of that KIR gene (42, 56).
Box 2
Alignment of full-length KIR cDNA sequences.
Exon–Intron Structure of the KIR Genes
Organization of the exon–intron structure of the various KIR genes is fairly consistent with the following basic arrangement: the signal sequence is encoded by the first two exons, each Ig domain (D0, D1, and D2, starting from the N-terminus) corresponds to a single exon (exons 3–5, respectively), the linker and transmembrane regions are each encoded by a single exon (exons 6 and 7), and the cytoplasmic domain is encoded by two final exons (Figure 3) (27, 57, 58). KIR2DL1 , 2DL2 /3 , and all 2DS genes [known as Type 1 two-domain KIR genes (59)] have an identical genomic organization to that encoding KIR molecules with three Ig domains. However, exon 3 is a pseudoexon in these two-domain KIR genes, which often remains in-frame but is eventually spliced out, possibly due to a three-base-pair deletion (59). The protein products of type 1 two-domain KIR are therefore missing the D0 domain (60). All NK cells express at least one type 1 2D KIR (61). The Type 2 two-domain KIR, which include 2DL4 , 2DL5A , and 2DL5B (59), are characterized by the complete absence of exon 4 (62), and therefore their protein product has no D1 domain. The 3DL3 gene closely resembles the other 3D genes, except that it is missing exon 6.
Two KIR pseudogenes have been identified and named, although others that closely resemble intact KIR genes (and therefore go undetected) may exist on certain haplotypes. KIR2DP1 (KIRZ) is closely related to 2DL2 /3 and 2DL1 (>97% homology at the nucleotide level), and contains two pseudoexons, 3 and 4. Pseudoexon 3 of 2DP1 contains the same aberrations as those identified in the Type 1 two-domain KIR genes, and a single base pair deletion in pseudoexon 4 of 2DP1 causes a frame shift that introduces a stop codon. A second KIR pseudogene, 3DP1 (KIRX), is severely truncated and alternate forms of the gene are differentiated by a 1.5kb deletion, which removes exon 2 (27). No transcripts for either 2DP1 or 3DP1 have been identified to date.
KIR Gene Order and Haplotypic Variability
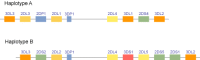
The KIRs are situated within a segment of DNA that has undergone expansion and contraction over time, and inspection of KIR haplotypes suggests a history of gene duplication and unequal crossing over in the region. The order of the KIR genes along the chromosome has been determined for two distinct haplotypes (Figure 4), providing a framework for their genomic order (27, 46, 63). The genes are organized in a head-to-tail fashion, and each gene is roughly 10–16 kb in length with a sequence of about 2 kb separating each pair of genes, except for a 14 kb stretch of unique sequence upstream of 2DL4 . Variation at the KIR gene complex is a function of both allelic polymorphism at several KIR genes and variability in the number and types of genes present on any given haplotype (3, 25, 64).
Although KIR haplotypes vary in the number and type of genes present (3, 65, 66, 70), the genes 2DL4 , 3DP1, 3DL2 , and 3DL3 are present on virtually all haplotypes and have therefore been termed framework loci (27). All others exist on only a fraction of the total haplotypic pool. The number of putatively expressed KIR genes present on a single haplotype ranges from about 7–12, depending primarily on the presence or absence of activating KIR loci (3, 27, 66). Based on gene content, the haplotypes have been divided into two primary sets, termed A and B, which were originally differentiated by the presence of a 24 kb HindIII fragment on Southern blot analysis (3). Haplotype A has seven loci; 2DL1 , 2DL3 , 2DL4 , 2DS4 , 3DL1 , 3DL2 and 3DL3 . Perhaps the most functionally relevant distinction between haplotypes A and B is the number of stimulatory receptors present. Haplotype A contains only a single stimulatory KIR gene, 2DS4 , whereas haplotype B contains various combinations of 2DS1 , 2DS2 , 2DS3 , 2DS5 , 3DS1 , and 2DS4 . Furthermore, the 2DS4 gene has a null allele with a population frequency of about 84% (allele frequency of 60%) (67) (and PN, unpublished observation). Thus, some individuals are homozygous for an A haplotype from which no activating KIR is expressed (68). The phenomenon of framework genes supporting areas of variable polygeny is analogous to that seen for HLA-DR, in which the DRA genes are always present, whereas the DRB gene number is variable (69).
The frequencies of haplotypes A and B are roughly equal in Caucasian populations, but on the basis of gene content, haplotype B displays a much greater variety of subtypes. Patterns of linkage disequilibrium (LD) between KIR loci (as opposed to LD between alleles of two genes that are both present on all haplotypes) are fixed for the A haplotypes (i.e. A haplotypes differ at the allelic level (25), but not in gene content). Much of what we know about LD patterns among the B haplotypes is based on studies of KIR profiles (presence or absence of each gene in a given individual) in groups of unrelated individuals, and they indicate that LD is quite strong between many pairs of genes represented in the group of B haplotypes (3, 65, 66, 70–72). Over 100 different KIR profiles have been identified among unrelated individuals, and many distinct gene-content haplotypes have been identified from segregation analysis (Figure 5) (68, 73, 74). The numbers will undoubtedly continue to grow as more individuals and families are screened.
Expansion and contraction of the KIR region appears to have occurred due in part to unequal crossing over. One consequence of such molecular genetic events is the possible generation of KIR haplotypes that have two (or more) copies of a gene on a single haplotype and the rearrangement of gene order. 2DL5 is the most recently identified KIR gene (75) and segregation analysis has indicated that what were once considered alleles of a single 2DL5 locus are actually two different loci, both of which can be present on a single haplotype (74, 76). Interestingly, these genes, termed 2DL5A and 2DL5B, are not tandemly located on haplotypes containing both genes, suggesting that they may have arisen due to a mechanism involving nonreciprocal crossing over. The two genes share >99% sequence similarity in both their exons and introns, and would not have been identified as separate loci if family studies of 2DL5 inheritance had not been performed (74, 76). There is at least one haplotype with several KIR deleted, including 2DL4 and 3DL1 (72). Since most KIR typing methods are designed to determine presence or absence of genes and subtyping of individual KIR genes in families has been performed in only a limited number of studies (25, 55, 77), haplotypes containing multiple copies of the same gene would remain undisclosed, further underestimating KIR haplotypic diversity.
Frequencies of specific KIR haplotypes and the two major haplotypic groups, A and B, vary across ethnically defined populations (65, 66, 70–72, 78–80). The A haplotype has an allele frequency of 75% in Japanese but only 15% in Australian Aborigines [estimated from (71, 79)]. The greatest intra-population haplotype diversity would appear to be in South Asians (72, 80) and the least in Japanese (79). It will be of interest to determine whether significant differences in KIR haplotypes across populations might account for variation in disease susceptibility among these groups.
Allelic Variability
Sequence analysis of KIR cDNA has shown that most KIR genes contain variable sites, and that some are quite polymorphic (25, 55, 64, 77, 81–91). Allelic polymorphism provides additional diversity to the extent that unrelated individuals identical for both KIR haplotypes are unlikely to be observed (25). The variation in KIR sequences can occur at positions encoding residues that affect interaction with HLA class I (92–95). Variation tends to occur throughout the gene, unlike the pattern observed in HLA class I and II genes where nucleotide variation is restricted primarily to one or two exons (96).
Similarity amongst KIR gene sequences and a history of unequal crossing over in the region has clouded the distinction between alleles of a single locus and separate gene loci. 2DS4 and 2DS1 were suggested to be allelic variants of a single locus (3), although more recent data suggests that they represent distinct loci (27, 65). 3DL1 and 3DS1 appear to occupy the same position on different haplotypes (27), and segregation analysis has indicated that they are indeed alleles of a single locus (55). We have previously proposed that 3DS1 , which is substantially less frequent than 3DL1 , arose by an unequal crossover between an ancestral 3DL1 gene and an ancestral activating KIR gene (6), based on the high sequence similarity between their extracellular domains and the observation that they segregate as alleles of a single gene. Nevertheless, rare haplotypes missing both or containing both 3DL1 and 3DS1 have been observed (55, 70). Haplotypes characterized by deletion of 3DL1 /3DS1 or by the presence of both 3DL1 and 3DS1 may have been derived from unequal crossovers that occurred subsequent to the event that formed 3DS1 . Wilson et al. (27) proposed that 2DL2 arose from a non-reciprocal recombination between 2DL1 and 2DL3 based on sequence similarity patterns, which would explain the observation that 2DL2 and 2DL3 also segregate as alleles of the same locus (65, 66, 70, 71). Moreover, the 2DP1 pseudogene, which is located between 2DL1 and 2DL3 and would have been lost in the cross over event, is indeed missing on 2DL2 haplotypes.
KIR3DL1 and 3DL2 , which encode molecules that bind certain allotypes of the HLA-B and HLA-A, are both quite polymorphic, although the source of variability in the two genes appears to be distinct (55). Recombination is likely to have generated much of the diversity in both genes (25). KIR3DL1 alleles encode molecules that appear to be expressed at different levels on the surface of NK cells based on antibody binding to 3DL1 allotypes. Allotypes with high, low and no binding have been observed and expression levels correlate with variation at specific amino acid residues of the 3DL1 molecule (55). It will be interesting to consider the functional consequences of these differences in expression patterns. Like the variability that is observed based on KIR gene content, allelic variability and frequencies also appear to distinguish different ethnic groups (83).
Linkage disequilibrium (LD) studies between pairs of polymorphic genes within the KIR locus are starting to emerge (25) revealing a pattern of strong allelic disequilibrium between pairs of genes located centromeric and pairs located telomeric of 2DL4 . Although significant in many instances, weaker disequilibrium patterns were observed between pairs of genes located in opposite halves of the locus. In general, patterns of LD that have been observed to date appear to correspond quite well with physical distance between genes.
Promoter Region Variability
The promoter regions of most KIR genes share >91% sequence similarity (26), and may therefore be controlled by similar mechanisms. The promoter regions of 3DL3 and 2DL4 , on the other hand, are more divergent (89% and 69% sequence similarity, respectively). Differences in promoter regions of these framework loci may account for the lack of 3DL3 expression and, alternatively, the expression of 2DL4 in virtually 100% of NK cell clones, a characteristic unique to 2DL4 (26). 2DL5A appears to be expressed based on measurements of mRNA in NK cells, but the most common allele of 2DL5B (in Caucasoids) is not expressed (76). Lack of 2DL5B expression correlates with a mutation in a putative AML1 transcription factor site in the promoter region, a variant that is also present in the pseudogene 3DP1 (76) and that may prevent expression of these two genes.
KIR Genotyping
KIR genotyping can be locus or allele specific. Locus only typing detects presence or absence of each gene in a given individual, thus providing a profile of the KIR repertoire (KIR profile). The PCR sequence-specific priming (PCR-SSP) method for KIR typing first described by Uhrberg and coworkers (3) has been updated to account for newly discovered loci and previously undetected alleles (65, 72, 78, 97, 98). A PCR sequence-specific oligonuleotide probe (PCR-SSOP) method has also been developed (70, 99). Inter-laboratory collaboration has helped to authenticate the molecular genotyping assay for KIR loci (100). Over 100 different KIR genotype profiles have been found so far; a summary of those published is shown in Figure 6.
Medium- to high-resolution allele-specific reactions (PCR-SSP) have been described for 2DL1 , 2DL3 , 3DL1 and 3DL2 (25, 55), and a single-stranded conformational polymorphism (SSCP) assay has been used to genotype 2DL4 (77, 91). Development of a comprehensive assay was required for 2DL5 in order to distinguish the two highly homologous loci, 2DL5A and 2DL5B (97). Reverse-transcriptase PCR (RT-PCR) based on PCR-SSP is the method of choice for allotyping NK cell clones and remains largely unchanged from that described previously (3). Various monoclonal antibodies are available for this purpose, but specificity is limited by the high homology between KIR isotypes.
Evolutionary Aspects
Comparisons of KIR sequences and haplotypes within and across species indicate that the KIR gene family is evolving rapidly, perhaps in response to species-specific pathogenic organisms (42, 101–103). It was initially thought that KIR were only present in higher primates, although KIR-like sequences have been found recently in lower primates (104, 105), ungulates (106) and other mammals (40). In chimpanzee, the closest living species to humans, ten KIR genes have been identified, only three of which appear to be direct orthologues of human KIR (42). Non-orthologous KIR genes have also been identified in pygmy chimpanzee (102), orangutan (101), rhesus monkeys (105), and baboon (104). The phylogenetic relationship of KIR genes from four different primate species is shown in Figure 7 [reproduced from (59); see also (101, 107)]. Primate species vary in ratios of long tail to short tail KIR genes, and haplotypic structure may also distinguish some species. For example, a common short haplotype of only three KIR genes distinctly characterizes pygmy chimpanzees (102). Comparisons of KIR gene sequences from five species of primates have revealed most lucidly the historical instability of the KIR genes in primate species. Nevertheless, similarities have persisted over millions of years, including the maintenance of 2DL4 in all primate species tested (although all 2DL4 transcripts examined in orangutan prematurely terminated). Further, identical receptor specificity for MHC-Cw molecules was observed in human and chimpanzees, and the two species are xenocompatible in that KIR receptors from chimpanzee can functionally recognize some human MHC class I molecules and vice versa.
Rapid evolution of the KIR locus may have resulted in species-specific characteristics across orders of mammals (108), potentially hampering attempts to clone KIR homologues in some species. KIR have now been detected in rodents (see; XM_142159, XM_142160, AF548540, AF548541, AY152727). Ly49 receptors, which belong to the C-type lectin domain family and are structurally unrelated to KIR, appear to perform the same function in rodents. Ly49 haplotypes are also complex, containing variable numbers of inhibitory and stimulatory genes (109), some of which are known to recognize mouse class I molecules. Genomes of humans and other primate species contain a single Ly49-like gene, which is a pseudogene in humans, gorilla, and chimpanzee (104, 110, 111), but may be functional in cow, baboon and orangutan (101, 104, 106).
It has been suggested that MHC class I and KIR (and also Ly49 in mouse) are coevolving (42, 112), such that as selection through infectious disease morbidity alters the frequencies and repertoire of class I variants, KIR/Ly49 must evolve to maintain or expand the ability to interact with class I in a beneficial manner. Evidence for the co-evolutionary process is illustrated by the observation that mouse class I allotypes lack determinants recognized by KIR, as do human class I allotypes for mouse Ly49 (86). Along these same lines, the orangutan have KIR2D genes that are predicted to encode receptors that specify only the Cw1 epitope (asparagine at position 80) of MHC-Cw molecules, correlating in an evolutionary sense with the observation that allotypes with the Cw2 epitope (lysine at position 80) are missing in this species and only MHC-Cw allotypes with the Cw1 epitope are available for interaction with 2D molecules (101). Additional selective pressures may also act directly on the KIR loci during early phases of infection by selecting for variants that enhance innate immune responsiveness, potentially increasing the rate of evolution at a speed surpassing that at the HLA class I loci (42, 113). Conforming to this hypothesis, all functional HLA class I genes have chimpanzee orthologues (114), but there are only three human-chimpanzee KIR orthologues (113). Thus, the portrayal of the KIR gene complex as the epitome of "eternal evolutionary restlessness" (58) appears to represent a precise account of the region.
Individual Molecular Characteristics
KIR2DL1
KIR2DL1 is a member of the type 1 (D1-D2) 2DL subfamily of inhibitory receptors. Exon 3 of 2DL1 is a pseudoexon that would otherwise encode the D0 domain. Ligands for 2DL1 are HLA-Cw molecules that have Asn77 and Lys80 (Cw*02/4/5/6/707/12042/15/1602/17). Monoclonal antibodies EB6 (CD158a) and HP3E4 recognize 2DL1, both of which also react with 2DS1 (115, 116). 2DL1*004 (2DL1v; inhibitory) appears to have been formed by a recombination between 2DL1 and 2DS1 .
Key references: 2DL1*004 (84); allelic variation (25); crystal structure of 2DL1 (94); crystal structure of 2DL1 contacting HLA-Cw4 (92).
Unique sequences:
PDB: 1NKR
LocusLink: 3802
OMIM: 604936
PROW: 2DL1.
KIR2DL2
KIR2DL2 is a member of the type 1 (D1-D2) 2DL subfamily of inhibitory receptors. Exon 3 of 2DL2 is a pseudoexon that would otherwise encode the D0 domain. Ligands for 2DL2 are HLA-Cw molecules that have Ser77 and Asn80 (Cw*01/3/7/8/12/13/14/1507/1601 and B*4601). Monoclonal antibodies GL183 (CD158b) and CH-L recognize 2DL2 as well as 2DL3 and 2DS2 (211, 212).
Key references: 2DL2 arose from a non-reciprocal recombination between 2DL1 and 2DL3 (27); 2DL2 and 2DL3 are alleles (66); crystal of 2DL2 (117); crystal structure of 2DL2 contacting HLA-Cw3 (1EFX) (93).
Unique sequences:
PDB: 1EFX
LocusLink: 3803
OMIM: 604937
PROW: 2DL2
KIR2DL3
KIR2DL3 is a member of the type 1 (D1-D2) 2DL subfamily of inhibitory receptors. Exon 3 of 2DL3 is a pseudoexon that would otherwise encode the D0 domain. Ligands for 2DL3 are HLA-Cw molecules that have Ser77 and Asn80 (Cw*01/3/7/8/12/13/14/1507/1601). Monoclonal antibodies GL183 (CD158b) and CH-L recognize 2DL3 as well as 2DL2 and 2DS2 (211, 212).
Key references: crystal structure of 2DL3 (118); 2DL3 and 2DL2 are alleles (66).
Unique sequences:
PDB: 1B6U
LocusLink: 3804
OMIM: 604938
PROW: 2DL3
KIR2DS1
KIR2DS1 is a type 1 (D1-D2) two-domain activating receptor. Exon 3 of 2DS1 is a pseudoexon that would otherwise encode the D0 domain. Ligands for 2DS1 may be HLA-Cw molecules that have Asn77 and Lys80 (Cw*02/4/5/6/707/12042/15/1602/17). Activating KIR signal by virtue of non-covalent association with the ITAM-bearing adaptor molecule, DAP-12. Monoclonal antibodies EB6 (CD158a) and HP3E4 recognize 2DS1, both of which also react with 2DL1 (115, 116).
Key references: 2DS1 associated with psoriatic arthritis when ligand for corresponding inhibitory receptor was absent (5).
Unique sequences:
LocusLink: 3806
OMIM: 604952
PROW: 2DS1
KIR2DS2
KIR2DS2 is a type 1(D1-D2) two-domain activating receptor. Exon 3 of 2DS2 is a pseudoexon that would otherwise encode the D0 domain. 2DS2 ligands are HLA-Cw molecules that have Ser77 and Asn80 (Cw*01/3/7/8/12/13/14/1507/1601). Activating KIR signal by virtue of non-covalent association with the ITAM-bearing adaptor molecule, DAP-12. Monoclonal antibodies GL183 (CD158b) and CH-L recognize 2DS2 as well as 2DL2 and 2DL3 (211, 212).
Key references: 2DS2 implicated in rheumatoid vasculitis (119); single substitution lowers binding affinity compared with 2DL2 (120); 2DS2 associated with psoriatic arthritis when ligand for corresponding inhibitory receptor was absent (5).
Unique sequences:
LocusLink: 3807
OMIM: 604953
PROW: 2DS2
KIR2DS3
KIR2DS3 is a type 1 (D1-D2) two-domain activating receptor. Exon 3 of 2DS3 is a pseudoexon that would otherwise encode the D0 domain. Its ligand is unknown. Activating KIR signal by virtue of non-covalent association with the ITAM-bearing adaptor molecule, DAP-12.
Unique sequences:
LocusLink: 3808
OMIM: 604954
PROW: 2DS3
KIR2DS4
KIR2DS4 is a type 1 (D1-D2) two-domain activating receptor. Exon 3 of 2DS4 is a pseudoexon that would otherwise encode the D0 domain. Ligand unknown. Activating KIR signal by virtue of non-covalent association with the ITAM-bearing adaptor molecule, DAP-12. Some alleles of 2DS4 have a 22bp deletion, which may lead to a truncated molecule (67), occasionally termed KIR1D (68).
Key references: Truncated 2DS4 allele (67); truncated 2DS4 on many A haplotypes (68); KIR2DS4 may bind HLA Cw4 (121).
Unique sequences:
LocusLink: 3809
OMIM: 604955
PROW: 2DS4
KIR2DS5
KIR2DS5 is a type 1 (D1-D2) two-domain activating receptor. Exon 3 of 2DS5 is a pseudoexon that would otherwise encode the D0 domain. Ligand unknown. Activating KIR signal by virtue of non-covalent association with the ITAM-bearing adaptor molecule, DAP-12.
Key references: (90)
Unique sequences:
- Pseudoexon 3: AF215833
LocusLink: 3810
OMIM: 604956
PROW: 2DS5
KIR2DL4
KIR2DL4 is a member of the type 2 (D0-D2) two-domain receptors. There is no exon 4 (D1). 2DL4 may transmit inhibitory, stimulatory, or both types of signals. 2DL4 probably binds to HLA-G. Some alleles of 2DL4 do not have a complete transmembrane domain but it is not clear whether these retain any function.
2DL4 is present on most haplotypes (a KIR framework locus).
Key references: 2DL4 binds to HLA-G (122, 123); or does it?(124); truncated alleles described (77); SNP (rs649216) (T) associated with truncated 2DL4 (91); 2DL4 is not always present (72); 2DL4 truncated in Orangutan (101); 2DL4 signaling (125, 126).
Unique sequences:
- 2DL4*001: X99480 (NK3.3)
- 2DL4*00201: X97229
- 2DL4*00202: AF034772
- 2DL4*003: AF003116 (KIR103)
- 2DL4*004: AF002979
- 2DL4*005: AF034771
- 2DL4*006: AF034773
- 2DL4*007: AF276292
LocusLink: 3805
OMIM: 604945
PROW: 2DL4
KIR2DL5
KIR2DL5 is a member of the type 2 (D0-D2) two-domain receptors. There is no exon 4 (D1). 2DL5 is likely to transmit inhibitory signals (Figure 2). Ligand is unknown. 2DL5 is the locus that caused the extra RFLP fragment reported by Uhrberg and coworkers and thus originally defined the KIR `B' haplotype (3, 59). There are two paralogous loci for this KIR. 2DL5A is in the telomeric KIR region and has one known allele, 2DL5A*001 (see below). 2DL5B is found in the centromeric KIR region and has three known alleles. 2DL5B*002 is not expressed.
Key references: 2DL5 characterized (75); 2DL5B*002 is not expressed (76); 2DL5 duplicated on some haplotypes (74).
Unique sequences:
LocusLink: 57292
OMIM: 605305
KIR3DL1
KIR3DL1 is a three-immunoglobulin-domain inhibitory receptor. One allele, 3DS1, is a three-immunoglobulin-domain activating receptor. 3DL1 interacts with HLA-B molecules that contain a Bw4 motif. Two commercially available clones, DX9 and Z27 react with 3DL1. DX9 and Z27 will bind to variants of 3DL1 with differing degrees of affinity; neither bind to 3DS1(55).
Key references: 3DL1 polymorphic(83); 3DL1 and 3DS1 alleles: high, low and no expression variants of 3DL1 (55); 3DS1 slows HIV disease progression if the correct HLA ligand is also present (6); 3DS1 rare in Africans (72); locus duplicated on some haplotypes (99).
Unique sequences:
- 3DL1*00102: AF262968
- 3DL1*003: AF022049
- 3DL1*00401: AF262969
- 3DL1*00402: AF262970
- 3DL1*005: AF262971
- 3DL1*006: AF262972
- 3DL1*007: AF262973
- 3DL1*008: AF262974
- 3DS1*004: AF022044
- 3DS1: AJ417558
- #3DS1*001: L76661
- #3DS1*002: X97233
- #3DS1*003: U73396
- 3DL1/3DL2 hybrid: AY059417
#not detected by PCR (55).
LocusLink: 3DL1: 3811; 3DS1: 3813
KIR3DL2
KIR3DL2 is a three-immunoglobulin-domain inhibitory receptor, which interacts with some HLA-A alleles. Monoclonal antibody DX31 binds to 3DL2.
3DL2 is present on most, if not all KIR haplotypes (a KIR framework locus).
Unique sequences:
- 3DL2*004: X93595
- 3DL2*005: L76666
- 3DL2*006: AF262966
- 3DL2*007: AF262965
- 3DL2*008: AF262967
- 3DL2*009: AF263617
- 3DL2*010: AY054918
- 3DL2*011: AY054919
- 3DL2*012: AY054920
- X93596 (NK18)
- 3DL1/3DL2 hybrid: AY059417
LocusLink: 3812
OMIM: 604947
PROW: 3DL2
KIR3DL3
KIR3DL3 is a three-immunoglobulin-domain receptor of unknown function or ligand. The 3DL3 gene closely resembles the other 3D genes, except that it is missing exon 6 (stalk region). 3DL3 is present on most, if not all KIR haplotypes (a KIR framework locus).
Unique sequences:
- AF352324 (3DL7)
LocusLink: 115653
KIR Biology
KIR Expression
KIR are expressed by classical NK and subpopulations of T cells (127, 128). NK clones from a single individual can vary substantially in the type of KIR molecules they express. Each cell requires at least one inhibitory receptor, such that when there is no appropriate inhibitory KIR–ligand combination, then other types of inhibitory receptor interactions will compensate (26, 129–131). With the exception of null alleles and possibly 3DL3 , all known KIR in a given individual's KIR gene repertoire are expressed, albeit apparently stochastically in order to generate a large number of different and overlapping subsets of KIR expression patterns (26, 132, 133). In heterozygous individuals, NK clones can express none, one or both alleles of 3DL1 and 3DL2 , but usually both alleles of 2DL4 (134). This somatic diversity means that there should be at least one NK clonal population that can respond to downregulation of any single HLA class I isotype (59, 135, 136).
KIR expression is likely to involve up to 15 different transcription promoter sequences which are located within 500 bp and 5' to the initiation codon (58). Other mechanisms controlling KIR levels are also apparent, including hormonal and cytokine regulation of expression (122, 137). Despite the many potential factors contributing to altered expression, long-term expression levels appear stable (131). The KIR locus maps to chromosome 19, whereas HLA maps to chromosome 6, and therefore the two loci segregate independently. Thus, NK cells can express KIR for which no known HLA ligand is present (129, 138, 139) and can also be expressed in individuals with very low HLA class I expression (140). Nevertheless, HLA can influence the number of peripheral blood KIR-expressing cells, but not the level of expression (131).
KIR Ligands
The ligands for several of the inhibitory KIR have been shown to be subsets of HLA class I molecules based on assays measuring binding of inhibitory KIR to specific HLA molecules and inhibition of NK-mediated cytolysis of target cells bearing those HLA allotypes [see (59, 141, 142)].
Dimorphisms in the HLA-Cw α1 domain that are characterized by Ser77/Asn80 and Asn77/Lys80 define serologically distinct allotypes of HLA-Cw (Cw group 1 and group 2, respectively). KIR2DL1 and 2DS1 interact with group 2 allotypes, while 2DL2, 2DL3 and 2DS2 interact with the alternative group 1 allotypes (143–145). The specificity for Cw type is defined entirely by a single substitution at KIR2D position 44 (145, 146). Importantly, NK cells expressing only Cw group 2 specific inhibitory receptors can lyse targets that are homozygous for the Cw group 1 allotypes and vice versa.
KIR3DL1 interacts with HLA-B allotypes that contain Bw4 (112, 147), a serologically-defined motif, and 3DL2 interacts with some HLA-A allotypes (87, 89). No KIR has yet been shown to bind allotypes containing Bw6, the alternative to Bw4 positive allotypes. Bw4 and Bw6 are also distinguished by polymorphism at positions 77 and 80. However, additional residues must be required for interaction with 3DL1, as some HLA-A allotypes possess the Bw4 motif but are not able to bind 3DL1 (112).
KIR2DL4, which is unusual by nature of its extracellular domain organization and the possession of only one ITIM, probably interacts with HLA-G (122, 123, 148). A summary of KIR isotypes and their specificity are shown in Table 3.
Table 3
KIR ligand specificity.
KIR Structure
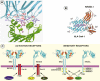
Crystal images of the two Ig domains for each of 2DL1 [1NKR; (94)], 2DL2 [1EFX; (117)] and 2DL3 [1B6U; (118)] are presently available. The ligand-contacting region falls at the membrane distal side of the junction between the Ig domains, which folds the D1 domain towards the cell surface. 2DL1–3 have similar structures but are distinguished by the angle between the Ig domains. This (hinge) angle appears to be stable (93), so morphological changes upon ligation are unlikely.
Type 1 2D KIR interact directly with the α-helices and bound peptide of their HLA ligands (92, 93). Ribbon diagrams of KIR-HLA interactions determined from crystal structures are shown in Figure 8. Six loops contact the MHC molecule, three from D1, one from the hinge and two from D2; all contact points result from complementary charges (93). There are also several hydrogen bonds, salt bridges and a degree of hydrophobic interaction. These high-resolution images also indicated that the KIR molecule contacts two residues from the bound peptide and forms a hydrogen bond with one of them (92, 93).
The D1 and D2 domains of three Ig domain KIR are likely to form a similar interface with Class I as those described for type I 2D KIR (120). Directed mutation experiments have shown that the D0 domain increases the avidity of this interaction (149), although all three Ig domains of 3DL1 were required for binding to HLA-B*51 (150).
KIR Interactions
KIR and TCR have overlapping footprints and therefore cannot bind to the same HLA/peptide complex concurrently (92, 93). This may impart some control over T cell activity over and above direct intracellular signaling, since KIR and TCR would be in competition for HLA ligand binding. The KIR\HLA interaction forms the basis of a natural killer immune synapse during immunosurveillance (151, 152), a process that has been illustrated by confocal microscopy (153). In these images, KIR can be seen inducing a ring formation of HLA-Cw molecules around a cluster of adhesion molecules at the cell surface. Receptor co-aggregation is likely to increase the signaling potential (117, 120). HLA and other membrane molecules may even be captured by the NK cell and either be internalized or remain at the NK cell surface (154, 155). The physiological role for intercellular transfer of HLA molecules remains to be determined, but may represent a novel mechanism for controlling NK cell activity.
Inhibitory KIR can actively prevent the localization of lipid rafts to the immune synapse (156, 157). Rafts contain complexes of activating accessory molecules, potentially explaining the dominant phenotype displayed by inhibitory receptors. The distinct binding affinities of activating compared to inhibitory KIR may also contribute to the dominance of inhibition. For example, 2DL2 has a higher affinity than 2DS2 for HLA-Cw3 due to a single amino acid substitution (120).
NK cells will become activated when inhibition is removed, so activation must involve stimulatory receptors (17). Based on assays measuring target cell killing, stimulatory KIR can mediate NK cell activity through recognition of HLA ligands (158), but little if any direct binding of activating KIR molecules to their putative HLA ligands has been detected and their high affinity, physiological ligands remain in question. Candidate ligands include non-MHC molecules, such as foreign or microbial antigens expressed on infected cells, normal cell surface proteins that are aberrantly expressed, stress-induced proteins or complexes of pathogen-derived peptides bound to MHC class I molecules. Recently, the mouse cytomegalovirus m157 gene product was shown to bind the mouse activating NK cell receptor Ly49H, an interaction that leads to NK cell killing of the infected targets (159–164). Although they lack sequence homology, the mouse Ly49 and human KIR families are considered to be functionally equivalent (165). Ly49H recognition of m157 provides strong support for the possibility that non-HLA molecules can behave as ligands for activating KIR.
As discussed under the section entitled KIR Ligands, specificity of inhibitory KIR for HLA-Cw allotypes is dictated to a large extent by the presence of asparagine or lysine at position 80 of the HLA-Cw molecule (115). Crystal structures supported this division of ligand specificity and suggested that the bound peptide also affects KIR binding, as there is a size limit to one of the KIR-contacting residues (92, 93). Although some interactions with KIR have been shown to be independent of peptide (166), several early studies also indicated that specificity depends on the presented peptide (167–171). Peptide-dependent protection of killing by NK cells has been observed (172, 173). Therefore, it has been suggested that inhibiting and activating receptors specific for the same HLA may respond differentially depending on bound peptides (138, 174). Peptide recognition could provide KIR with one further means for mediating pathogen-specific immunity.
KIR Signaling
The exact mechanism for signal transmission to KIR transmembrane domains has not been elucidated. Most inhibitory KIR have two ITIMs (Box 2) and these operate in tandem to mediate inhibition (175, 176). ITIM phosphorylation results in association with SHP (Src-homology domain-bearing tyrosine phosphatase), which specifically inhibits the proteins involved in the intracellular activation cascade (177). Activating KIR have no direct signaling properties but associate with the ITAM-bearing adaptor molecule, DAP12. DAP12 and short-tailed KIR associate due to complementarily charged transmembrane residues (52, 53). Ligation of activating KIR leads to phosphorylation of DAP12, recruitment of ZAP-70/Syk kinase and the induction of an intracellular signaling cascade (178, 179). Also, ligation of activating KIR may control the response by inducing apoptosis of mature cytotoxic cells (180).
One exception to the pattern of division described for stimulatory and inhibitory receptors is 2DL4 (123, 148, 181), which has a long cytoplasmic tail containing only a single ITIM motif. 2DL4 also contains a charged arginine residue in the transmembrane region, which may facilitate interaction with DAP12. Thus, it has been proposed that 2DL4 may transmit inhibitory, stimulatory, or both types of signals (125, 126). Some alleles of 2DL4 do not have a complete transmembrane domain (77), but it is not clear whether these are expressed at the cell surface or retain any function.
NK Development
Classical NK cells develop from CD34pos progenitor cells in the bone marrow, in the presence of stromally derived cytokines such as IL15. KIR may be involved in this development (182), suggesting some form of selection for the NK cell receptor repertoire. A sequential pattern of KIR expression during NK maturation may occur, which could ensure self tolerance (133, 183). Clonal NK cell expression of a subset of KIR is stable over time and is predominantly determined by KIR genotype, with only moderate influence from HLA (131), if at all (182). KIR promoter methylation correlates with stable KIR expression, but it is not clear at which stage in development methylation patterns are established (134, 184). As KIR can be expressed for which there is no known HLA ligand and vice-versa (129, 138), there appears to be no mechanism for maturation of NK cell clones through selection. The number of functional HLA genes, for example, is controlled to some extent by selection processes in the thymus, where too many HLA molecules would restrict the TCR repertoire and thus reduce immune potential (185). Thus, the constraint placed on MHC polygeny may not apply to KIR, potentially explaining the large and varying number of KIR genes.
CD56 Expression
Also known as N-CAM (neural cell adhesion molecule), CD56 is expressed on all NK as well as subpopulations of T cells (186), but its function remains elusive. NK cells expressing low levels of CD56 (CD56dim) tend to have more resting cytotoxic activity than those with high levels (CD56bri) (13). CD56dim NK cells express KIR, whereas CD56bri cells do not (187). It is unclear whether CD56 (and KIR) expression on a given NK cell changes with activation state (188) or if two developmental lineages occur (13). Regardless, these observations indicate that KIR expression is intrinsically linked to the cytotoxic potential of an immune cell. CD56bri cells are scarce in the periphery but have been found in lymph nodes, where they may play a role in the adaptive immune response (189).
Disease Phenotypes and Molecular Medicine
KIR in Adaptive Immunity
KIR can be expressed by αβ- and γδ- T cells, and they appear to be characteristic of memory CD8+ T cells, through which they may modify the response to antigenic re-challenge (137, 190, 191). KIR expression appears to be inducible during early clonal expansion of T cells (192). KIR may also be important in the transition to a memory phenotype (183, 193). As the spectrum of response occurs during infection, including self-reactivity, the capacity for reactivation must be controlled. Inhibition of T cell activity by KIR may ensure that a secondary response only occurs when the danger is sufficiently strong. Moreover, expression of inhibitory KIR by T cells during an active response may help cytolytic T cells to remain focused on infected cells (137). T cells that acquire NK activating receptors may be harmful if they do not express appropriate inhibitory KIR (194). Conversely, T cells aberrantly expressing inhibitory KIR may not remove disease cells effectively (137).
KIR and Disease
Only a limited number of studies addressing genetic associations between KIR genes and specific diseases have been reported to date, primarily due to the very recent and ongoing characterization of the genes and their haplotypes. Natural killer cells have been implicated in the defense against infectious diseases, particularly viral infections, through mechanisms involving cytotoxicity and cytokine production (10), presumably mediated in part by stimulatory KIR molecules. Given the receptor–ligand relationship between certain combinations of KIR and HLA class I molecules, it is reasonable to hypothesize a synergistic relationship between these polymorphic loci that may ultimately regulate NK cell mediated immunity against infectious pathogens. Recently, we showed that 3DS1 in combination with HLA-B alleles that encode molecules with isoleucine at position 80 (HLA-B Bw4-80Ile) resulted in delayed progression to AIDS after HIV infection (6). An additive genetic effect of the two loci was deemed unlikely on the basis that HLA-B Bw4-80Ile alleles in the absence of 3DS1 were not associated with AIDS progression, and 3DS1 in absence of HLA-B Bw4-80Ile was significantly associated with more rapid progression to AIDS. Thus, a model involving a protective epistatic interaction between 3DS1 and HLA-B Bw4-80Ile was proposed whereby 3DS1 receptors bind HLA-B Bw4-80Ile (perhaps when they contain HIV peptides), leading to NK cell activation and elimination of HIV-1 infected cells.
Natural selection for resistance to a pathogen can lead to the increase in frequency of alleles that are otherwise deleterious. The extensive diversity of KIR haplotypes might suggest the possibility of pleiotropic KIR effects on different diseases, whereby a KIR gene conferring protection against one disease may predispose to another, perhaps less deadly disease. Two studies have implicated stimulatory KIR genes in increased risk of developing specific autoimmune diseases (5, 119). In the first study, 2DS2 was significantly more prevalent amongst patients with rheumatoid vasculitis compared to either normal individuals or patients with rheumatoid arthritis, but no vasculitis (119, 194). CD4+CD28null T cells, as opposed to NK cells, that expressed 2DS2 were implicated in vascular damage possibly due to stimulatory effects of 2DS2. The second study reported increased susceptibility to developing psoriatic arthritis amongst individuals with 2DS1 and/or 2DS2, but only when HLA ligand for their homologous inhibitory receptors, 2DL1 and 2DL2 /3, were missing (5). The data suggested that absence of ligands for inhibitory KIRs could potentially lower the threshold for NK (and/or T) cell activation mediated through stimulatory receptors, thereby contributing to disease pathogenesis.
Genome-wide linkage studies of celiac disease have suggested that candidate susceptibility loci map to the region of 19q13.4 (5, 195). Using association analyses, Moodie et al. (196) showed no association between celiac disease and KIR haplotypes distinguished by the presence or absence of 2DL1 , 2DL2 , 2DL3 and 2DL5 that could account for the linkage studies. However, this does not rule out allelic variation at any of the loci.
2DL4 binds to HLA-G (122, 123), a nonclassical class I molecule that is expressed on the human trophoblast (197), and this receptor-ligand interaction may confer some protection against maternal NK or T cell-mediated rejection of the hemi-allogeneic fetus. Several alleles of 2DL4 have been identified, some of which are characterized by a single nucleotide deletion that results in the elimination of exon 6 during mRNA production (77), raising the possibility that 2DL4 alleles may vary in their ability to control fetus rejection. However, a study of 2DL4 allelic variation and KIR gene frequencies in 45 women who experienced pre-eclampsia compared to 48 normotensive controls indicated no significant differences between the two groups (91).
Recently, KIR molecules have been implicated in reduced risk of relapse in patients with acute myeloid leukemia (AML) who received hematopoietic transplants that were mismatched for KIR ligands (7, 198). KIR ligand incompatibility was defined as absence in recipients of donor class I allelic groups known to be ligands for inhibitory KIRs (3DL1, 2DL2 /3, 2DL1 and 3DL2). The data indicated that donor-versus-recipient NK cell alloreactivity was capable of eliminating leukemia relapse and graft rejection, and it also protected patients against graft-versus-host-disease.
KIR binding specificity for groups of HLA molecules raises the possibility that mechanisms responsible for HLA associations with some diseases may stem from interactions with KIR molecules on NK or T cells, and a limited number of studies have lent strong support to this hypothesis. Further delineation of the diversity at the KIR gene complex and development of efficient typing systems will surely foster disease association studies that are as plentiful and varied as those that have addressed the HLA loci.
Acknowledgments
We are very grateful to Gary Smythers and Pat Martin for alignment of the KIR cDNA sequences.
We would also like to thank Robert Vaughan and South Thames Clinical Transplantation Laboratory for support during the preparation of this article.
Killer Reviews
Structure
Salter, R. D., H. W. Chan, R. Tadikamalla, and D. A. Lawlor. 1997. Domain organization and sequence relationship of killer cell inhibitory receptors. Immunol Rev 155:175 (199)
Boyington, J. C., A. G. Brooks, and P. D. Sun. 2001. Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol Rev 181:66 (120)
McCann, F. E., K. Suhling, L. M. Carlin, K. Eleme, S. B. Taner, K. Yanagi, B. Vanherberghen, P. M. French, and D. M. Davis. 2002. Imaging immune surveillance by T cells and NK cells. Immunol Rev 189:179 (200)
Vyas, Y. M., H. Maniar, and B. Dupont. 2002. Visualization of signaling pathways and cortical cytoskeleton in cytolytic and noncytolytic natural killer cell immune synapses. Immunol Rev 189:161 (201)
Function
Lanier, L. L. 1998. NK cell receptors. Annu Rev Immunol 16:359 (202)
Long, E. O. 1999. Regulation of immune responses through inhibitory receptors. Annu Rev Immunol 17:875 (190)
Biassoni, R., C. Cantoni, D. Pende, S. Sivori, S. Parolini, M. Vitale, C. Bottino, and A. Moretta. 2001. Human natural killer cell receptors and co-receptors. Immunol Rev 181:203 (203)
Moretta, A., C. Bottino, M. Vitale, D. Pende, C. Cantoni, M. C. Mingari, R. Biassoni, and L. Moretta. 2001. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol 19:197 (204)
Raulet, D. H., R. E. Vance, and C. W. McMahon. 2001. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol 19:291 (130)
Vilches, C., and P. Parham. 2002. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217 (59)
McQueen, K. L., and P. Parham. 2002. Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol 14:615 (205)
Genetics
Trowsdale, J., R. Barten, A. Haude, C. A. Stewart, S. Beck, and M. J. Wilson. 2001. The genomic context of natural killer receptor extended gene families. Immunol Rev 181:20 (58)
Trowsdale, J. 2001. Genetic and functional relationships between MHC and NK receptor genes. Immunity 15:363 (206)
Vilches, C., and P. Parham. 2002. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217 (59)
Yawata, M. et al. 2003. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol 22:463-482 (214)
References
- 1.
- Harel-Bellan A , Quillet A , Marchiol C , DeMars R , Tursz T , Fradelizi D . Natural killer susceptibility of human cells may be regulated by genes in the HLA region on chromosome 6. Proc Natl Acad Sci U S A. 1986;83:5688. [PMC free article: PMC386354] [PubMed: 2426704]
- 2.
- Moretta A , Tambussi G , Bottino C C , Tripodi G , Merli A , Ciccone E , Pantaleo G , Moretta L . A novel surface antigen expressed by a subset of human CD3- CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J Exp Med. 1990;171:695. [PMC free article: PMC2187781] [PubMed: 2137855]
- 3.
- Uhrberg M , Valiante N M , Shum B P , Shilling H G , Lienert-Weidenbach K , Corliss B , Tyan D , Lanier L L , Parham P . Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753. [PubMed: 9430221]
- 4.
- Warrington K J , Takemura S , Goronzy J J , Weyand C M . CD4+,CD28- T cells in rheumatoid arthritis patients combine features of the innate and adaptive immune systems. Arthritis Rheum. 2001;44:13. [PubMed: 11212151]
- 5.
- Martin M P , Nelson G , Lee J H , Pellett F , Gao X , Wade J , Wilson M J , Trowsdale J , Gladman D , Carrington M . Cutting Edge: Susceptibility to Psoriatic Arthritis: Influence of Activating Killer Ig-Like Receptor Genes in the Absence of Specific HLA- C Alleles. J Immunol. 2002;169:2818. [PubMed: 12218090]
- 6.
- Martin M P , Gao X , Lee J H , Nelson G W , Detels R , Goedert J J , Buchbinder S , Hoots K , Vlahov D , Trowsdale J , Wilson M , O'Brien S J , Carrington M . Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429. [PubMed: 12134147]
- 7.
- Ruggeri L , Capanni M , Urbani E , Perruccio K , Shlomchik W D , Tosti A , Posati S , Rogaia D , Frassoni F , Aversa F , Martelli M F , Velardi A . Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097. [PubMed: 11896281]
- 8.
- Shilling H , McQueen K L , Cheng N W , Shizuru J A , Negrin R S , Parham P . Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101:3730–3740. [PubMed: 12511415]
- 9.
- Bancroft G J . The role of natural killer cells in innate resistance to infection. Curr Opin Immunol. 1993;5:503. [PubMed: 8216925]
- 10.
- Biron C A , Nguyen K B , Pien G C , Cousens L P , Salazar-Mather T P . Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189. [PubMed: 10358757]
- 11.
- Robertson M J , Ritz J . Biology and clinical relevance of human natural killer cells. Blood. 1990;76:2421. [PubMed: 2265240]
- 12.
- Trinchieri G . Biology of natural killer cells. Adv Immunol. 1989;47:187. [PMC free article: PMC7131425] [PubMed: 2683611]
- 13.
- Cooper M A , Fehniger T A , Caligiuri M A . The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633. [PubMed: 11698225]
- 14.
- Bjorkman P J , Saper M A , Samraoui B , Bennett W S , Strominger J L , Wiley D C . The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512. [PubMed: 2443855]
- 15.
- Zinkernagel R M , Doherty P C . Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701. [PubMed: 4133807]
- 16.
- Storkus W J , Alexander J , Payne J A , Dawson J R , Cresswell P . Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc Natl Acad Sci U S A. 1989;86:2361. [PMC free article: PMC286912] [PubMed: 2784569]
- 17.
- Karre K , Ljunggren H G , Piontek G , Kiessling R . Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675. [PubMed: 3951539]
- 18.
- Shimizu Y , DeMars R . Demonstration by class I gene transfer that reduced susceptibility of human cells to natural killer cell-mediated lysis is inversely correlated with HLA class I antigen expression. Eur J Immunol. 1989;19:447. [PubMed: 2707298]
- 19.
- Ljunggren H G , Karre K . In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990;11:237. [PubMed: 2201309]
- 20.
- Biassoni R , Pessino A , Malaspina A , Cantoni C , Bottino C , Sivori S , Moretta L , Moretta A . Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095. [PubMed: 9464792]
- 21.
- Sivori S , Vitale M , Morelli L , Sanseverino L , Augugliaro R , Bottino C , Moretta L , Moretta A . p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med. 1997;186:1129. [PMC free article: PMC2211712] [PubMed: 9314561]
- 22.
- Vales-Gomez M , Reyburn H T , Erskine R A , Strominger J . Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci U S A. 1998;95:14326. [PMC free article: PMC24372] [PubMed: 9826699]
- 23.
- Watzl C , Stebbins C C , Long E O . NK cell inhibitory receptors prevent tyrosine phosphorylation of the activation receptor 2B4 (CD244). J Immunol. 2000;165:3545. [PubMed: 11034353]
- 24.
- Falco M , Biassoni R , Bottino C , Vitale M , Sivori S , Augugliaro R , Moretta L , Moretta A . Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J Exp Med. 1999;190:793. [PMC free article: PMC2195632] [PubMed: 10499918]
- 25.
- Shilling H G , Guethlein L A , Cheng N W , Gardiner C M , Rodriguez R , Tyan D , Parham P . Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307. [PubMed: 11859120]
- 26.
- Valiante N M , Uhrberg M , Shilling H G , Lienert-Weidenbach K , Arnett K L , D'Andrea A , Phillips J H , Lanier L L , Parham P . Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739. [PubMed: 9430220]
- 27.
- Wilson M J , Torkar M , Haude A , Milne S , Jones T , Sheer D , Beck S , Trowsdale J . Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778. [PMC free article: PMC18309] [PubMed: 10781084]
- 28.
- Samaridis J , Colonna M . Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: structural evidence for new stimulatory and inhibitory pathways. Eur J Immunol. 1997;27:660. [PubMed: 9079806]
- 29.
- Cosman D , Fanger N , Borges L , Kubin M , Chin W , Peterson L , Hsu M L . A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273. [PubMed: 9285411]
- 30.
- Wagtmann N , Rojo S , Eichler E , Mohrenweiser H , Long E O . A new human gene complex encoding the killer cell inhibitory receptors and related monocyte/macrophage receptors. Curr Biol. 1997;7:615. [PubMed: 9259559]
- 31.
- Arm J P , Nwankwo C , Austen K F . Molecular identification of a novel family of human Ig superfamily members that possess immunoreceptor tyrosine-based inhibition motifs and homology to the mouse gp49B1 inhibitory receptor. J Immunol. 1997;159:2342. [PubMed: 9278324]
- 32.
- Colonna M . Immunology. Unmasking the killer's accomplice. Nature. 1998;391:642. [PubMed: 9490407]
- 33.
- Lepin E J , Bastin J M , Allan D S , Roncador G , Braud V M , Mason D Y , van der Merwe P A , McMichael A J , Bell J I , Powis S H , O'Callaghan C A . Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur J Immunol. 2000;30:3552. [PubMed: 11169396]
- 34.
- Vitale M , Castriconi R , Parolini S , Pende D , Hsu M L , Moretta L , Cosman D , Moretta A . The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: analysis of LIR-1 + NK cell clones. Int Immunol. 1999;11:29. [PubMed: 10050671]
- 35.
- Fanger N A , Borges L , Cosman D . The leukocyte immunoglobulin-like receptors (LIRs): a new family of immune regulators. J Leukoc Biol. 1999;66:231. [PubMed: 10449159]
- 36.
- Nakajima H , Cella M , Langen H , Friedlein A , Colonna M . Activating interactions in human NK cell recognition: the role of 2B4- CD48. Eur J Immunol. 1999;29:1676. [PubMed: 10359122]
- 37.
- Cella M , Dohring C , Samaridis J , Dessing M , Brockhaus M , Lanzavecchia A , Colonna M . A novel inhibitory receptor (ILT3) expressed on monocytes, macrophages, and dendritic cells involved in antigen processing. J Exp Med. 1997;185:1743. [PMC free article: PMC2196312] [PubMed: 9151699]
- 38.
- Canavez F , Young N T , Guethlein L A , Rajalingam R , Khakoo S I , Shum B P , Parham P . Comparison of chimpanzee and human leukocyte Ig-like receptor genes reveals framework and rapidly evolving genes. J Immunol. 2001;167:5786. [PubMed: 11698452]
- 39.
- Dennis G Jr.,, Kubagawa H , Cooper M D . Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc Natl Acad Sci U S A. 2000;97:13245. [PMC free article: PMC27210] [PubMed: 11078516]
- 40.
- Volz A , Wende H , Laun K , Ziegler A . Genesis of the ILT/LIR/MIR clusters within the human leukocyte receptor complex. Immunol Rev. 2001;181:39. [PubMed: 11513150]
- 41.
- Young N T , Canavez F , Uhrberg M , Shum B P , Parham P . Conserved organization of the ILT/LIR gene family within the polymorphic human leukocyte receptor complex. Immunogenetics. 2001;53:270. [PubMed: 11491530]
- 42.
- Khakoo S I , Rajalingam R , Shum B P , Weidenbach K , Flodin L , Muir D G , Canavez F , Cooper S L , Valiante N M , Lanier L L , Parham P . Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687. [PubMed: 10894168]
- 43.
- Vivier E , Daeron M . Immunoreceptor tyrosine-based inhibition motifs. Immunol Today. 1997;18:286. [PubMed: 9190115]
- 44.
- Colonna M , Nakajima H , Navarro F , Lopez-Botet M . A novel family of Ig-like receptors for HLA class I molecules that modulate function of lymphoid and myeloid cells. J Leukoc Biol. 1999;66:375. [PubMed: 10496306]
- 45.
- Torkar M , Norgate Z , Colonna M , Trowsdale J , Wilson M J . Isotypic variation of novel immunoglobulin-like transcript/killer cell inhibitory receptor loci in the leukocyte receptor complex. Eur J Immunol. 1998;28:3959. [PubMed: 9862332]
- 46.
- Wende H , Volz A , Ziegler A . Extensive gene duplications and a large inversion characterize the human leukocyte receptor cluster. Immunogenetics. 2000;51:703. [PubMed: 10941842]
- 47.
- Meyaard L , van der Vuurst de Vries A R , de Ruiter T , Lanier L L , Phillips J H , Clevers H . The epithelial cellular adhesion molecule (Ep-CAM) is a ligand for the leukocyte-associated immunoglobulin-like receptor (LAIR). J Exp Med. 2001;194:107. [PMC free article: PMC2193444] [PubMed: 11435477]
- 48.
- Colonna M , Navarro F , Bellon T , Llano M , Garcia P , Samaridis J , Angman L , Cella M , Lopez-Botet M . A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809. [PMC free article: PMC2199153] [PubMed: 9382880]
- 49.
- Norman P J , Carey B S , Stephens H A , Vaughan R W . DNA sequence variation and molecular genotyping of natural killer leukocyte immunoglobulin-like receptor, LILRA3. Immunogenetics. 2003 [PubMed: 12750859]
- 50.
- Andre P , Biassoni R , Colonna M , Cosman D , Lanier L L , Long E O , Lopez-Botet M , Moretta A , Moretta L , Parham P , Trowsdale J , Vivier E , Wagtmann N , Wilson M J . New nomenclature for MHC receptors. Nat Immunol. 2001;2:661. [PubMed: 11477395]
- 51.
- Hsu K C , Chida S , Dupont B , Geraghty D E . The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40. [PubMed: 12493005]
- 52.
- Olcese L , Cambiaggi A , Semenzato G , Bottino C , Moretta A , Vivier E . Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells. J Immunol. 1997;158:5083. [PubMed: 9164921]
- 53.
- Lanier L L , Corliss B C , Wu J , Leong C , Phillips J H . Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703. [PubMed: 9490415]
- 54.
- Blery M , Olcese L , Vivier E . Early signaling via inhibitory and activating NK receptors. Hum Immunol. 2000;61:51. [PubMed: 10658978]
- 55.
- Gardiner C M , Guethlein L A , Shilling H G , Pando M , Carr W H , Rajalingam R , Vilches C , Parham P . Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992. [PubMed: 11207248]
- 56.
- Steffens U , Vyas Y , Dupont B , Selvakumar A . Nucleotide and amino acid sequence alignment for human killer cell inhibitory receptors (KIR), 1998. Tissue Antigens. 1998;51:398. [PubMed: 9583814]
- 57.
- Wilson M J , Torkar M , Trowsdale J . Genomic organization of a human killer cell inhibitory receptor gene. Tissue Antigens. 1997;49:574. [PubMed: 9234478]
- 58.
- Trowsdale J , Barten R , Haude A , Stewart C A , Beck S , Wilson M J . The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20. [PubMed: 11513141]
- 59.
- Vilches C , Parham P . KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217. [PubMed: 11861603]
- 60.
- Vilches C , Pando M J , Parham P . Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000;51:639. [PubMed: 10941835]
- 61.
- Watzl C , Peterson M , Long E O . Homogenous expression of killer cell immunoglobulin-like receptors (KIR) on polyclonal natural killer cells detected by a monoclonal antibody to KIR2D. Tissue Antigens. 2000;56:240. [PubMed: 11034560]
- 62.
- Selvakumar A , Steffens U , Palanisamy N , Chaganti R S , Dupont B . Genomic organization and allelic polymorphism of the human killer cell inhibitory receptor gene KIR103. Tissue Antigens. 1997;49:564. [PubMed: 9234477]
- 63.
- Wende H , Colonna M , Ziegler A , Volz A . Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154. [PubMed: 9922396]
- 64.
- Selvakumar A , Steffens U , Dupont B . Polymorphism and domain variability of human killer cell inhibitory receptors. Immunol Rev. 1997;155:183. [PubMed: 9059894]
- 65.
- Norman P J , Stephens H A , Verity D H , Chandanayingyong D , Vaughan R W . Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52:195. [PubMed: 11220621]
- 66.
- Witt C S , Dewing C , Sayer D C , Uhrberg M , Parham P , Christiansen F T . Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. 1999;68:1784. [PubMed: 10609957]
- 67.
- Maxwell L D , Wallace A , Middleton D , Curran M D . A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens. 2002;60:254. [PubMed: 12445308]
- 68.
- Hsu K C , Liu X R , Selvakumar A , Mickelson E , O'Reilly R J , Dupont B . Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118. [PubMed: 12391228]
- 69.
- Campbell R D , Trowsdale J . Map of the human MHC. Immunol Today. 1993;14:349. [PubMed: 8363724]
- 70.
- Crum K A , Logue S E , Curran M D , Middleton D . Development of a PCR-SSOP approach capable of defining the natural killer cell inhibitory receptor (KIR) gene sequence repertoires. Tissue Antigens. 2000;56:313. [PubMed: 11098931]
- 71.
- Toneva M , Lepage V , Lafay G , Dulphy N , Busson M , Lester S , Vu-Trien A , Michaylova A , Naumova E , McCluskey J , Charron D . Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001;57:358. [PubMed: 11380947]
- 72.
- Norman P J , Carrington C V , Byng M , Maxwell L D , Curran M D , Stephens H A , Chandanayingyong D , Verity D H , Hameed K , Ramdath D D , Vaughan R W . Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun. 2002;3:86. [PubMed: 11960306]
- 73.
- Uhrberg M , Parham P , Wernet P . Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54:221. [PubMed: 12136333]
- 74.
- Gomez-Lozano N , Gardiner C M , Parham P , Vilches C . Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314. [PubMed: 12185535]
- 75.
- Vilches C , Rajalingam R , Uhrberg M , Gardiner C M , Young N T , Parham P . KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig- like domains. J Immunol. 2000;164:5797. [PubMed: 10820258]
- 76.
- Vilches C , Gardiner C M , Parham P . Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5 gene. J Immunol. 2000;165:6416. [PubMed: 11086080]
- 77.
- Witt C S , Martin A , Christiansen F T . Detection of KIR2DL4 alleles by sequencing and SSCP reveals a common allele with a shortened cytoplasmic tail. Tissue Antigens. 2000;56:248. [PubMed: 11034561]
- 78.
- Cook, M. A., P. A. H. Moss, and D. C. Briggs. 2003. The Distribution of 13 KIR loci in UK Blood Donors from 3 Ethnic Groups. Eur J Immunogenet In Press. [PubMed: 12787000]
- 79.
- Yawata M , Yawata N , McQueen K L , Cheng N W , Guethlein L A , Rajalingam R , Shilling H G , Parham P . Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54:543. [PubMed: 12439616]
- 80.
- Rajalingam R , Krausa P , Shilling H G , Stein J B , Balamurugan A , McGinnis M D , Cheng N W , Mehra N K , Parham P . Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics. 2002;53:1009. [PubMed: 11904677]
- 81.
- Colonna M , Samaridis J . Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405. [PubMed: 7716543]
- 82.
- Wagtmann N , Biassoni R , Cantoni C , Verdiani S , Malnati M S , Vitale M , Bottino C , Moretta L , Moretta A , Long E O . Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439. [PubMed: 7749980]
- 83.
- Rajalingam R , Gardiner C M , Canavez F , Vilches C , Parham P . Identification of seventeen novel KIR variants: fourteen of them from two non-Caucasian donors. Tissue Antigens. 2001;57:22. [PubMed: 11169255]
- 84.
- Shilling H G , Lienert-Weidenbach K , Valiante N M , Uhrberg M , Parham P . Evidence for recombination as a mechanism for KIR diversification. Immunogenetics. 1998;48:413. [PubMed: 9799338]
- 85.
- Dohring C , Samaridis J , Colonna M . Alternatively spliced forms of human killer inhibitory receptors. Immunogenetics. 1996;44:227. [PubMed: 8662091]
- 86.
- Valiante N M , Lienert K , Shilling H G , Smits B J , Parham P . Killer cell receptors: keeping pace with MHC class I evolution. Immunol Rev. 1997;155:155. [PubMed: 9059891]
- 87.
- Pende D , Biassoni R , Cantoni C , Verdiani S , Falco M , di Donato C , Accame L , Bottino C , Moretta A , Moretta L . The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996;184:505. [PMC free article: PMC2192700] [PubMed: 8760804]
- 88.
- D'Andrea A , Chang C , Franz-Bacon K , McClanahan T , Phillips J H , Lanier L L . Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J Immunol. 1995;155:2306. [PubMed: 7650366]
- 89.
- Dohring C , Scheidegger D , Samaridis J , Cella M , Colonna M . A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156:3098. [PubMed: 8617928]
- 90.
- Vilches C , Pando M J , Rajalingam R , Gardiner C M , Parham P . Discovery of two novel variants of KIR2DS5 reveals this gene to be a common component of human KIR 'B' haplotypes. Tissue Antigens. 2000;56:453. [PubMed: 11144295]
- 91.
- Witt C S , Whiteway J M , Warren H S , Barden A , Rogers M , Martin A , Beilin L , Christiansen F T . Alleles of the KIR2DL4 receptor and their lack of association with pre-eclampsia. Eur J Immunol. 2002;32:18. [PubMed: 11754000]
- 92.
- Fan Q R , Long E O , Wiley D C . Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452. [PubMed: 11323700]
- 93.
- Boyington J C , Motyka S A , Schuck P , Brooks A G , Sun P D . Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537. [PubMed: 10850706]
- 94.
- Fan Q R , Mosyak L , Winter C C , Wagtmann N , Long E O , Wiley D C . Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature. 1997;389:96. [PubMed: 9288975]
- 95.
- Boyington J C , Sun P D . A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007. [PubMed: 11955593]
- 96.
- Hughes A L . Natural selection and the diversification of vertebrate immune effectors. Immunol Rev. 2002;190:161. [PubMed: 12493013]
- 97.
- Gomez-Lozano N , Vilches C . Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 2002;59:184. [PubMed: 12074708]
- 98.
- Gagne K , Brizard G , Gueglio B , Milpied N , Herry P , Bonneville F , Cheneau M L , Schleinitz N , Cesbron A , Follea G , Harrousseau J L , Bignon J D . Relevance of KIR gene polymorphisms in bone marrow transplantation outcome. Hum Immunol. 2002;63:271. [PubMed: 12039408]
- 99.
- Maxwell, L. D., A. Meenagh, F. Williams, E. du Toit, D. Taljaard, A. Wallace, M. D. Curran, and D. Middleton. 2003. Definition of common KIR haplotypes in the human population using an improved KIR PCR-SSOP typing scheme and the characterisation of novel KIR variants. In XIII International Congress of Histocompatibility and Immunogenetics, Vol. In Press. Munksgaard.
- 100.
- Cook, M. A., P. J. Norman, M. D. Curran, L. D. Maxwell, D. C. Briggs, R. W. Vaughan, and D. Middleton. 2003. A Multi-Laboratory Characterisation of the KIR Genotypes of 10th International Histocompatibility Workshop Cell-lines. Hum Immunol in press. [PubMed: 12691708]
- 101.
- Guethlein L A , Flodin L R , Adams E J , Parham P . NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC- C. J Immunol. 2002;169:220. [PubMed: 12077248]
- 102.
- Rajalingam R , Hong M , Adams E J , Shum B P , Guethlein L A , Parham P . Short KIR haplotypes in pygmy chimpanzee (Bonobo) resemble the conserved framework of diverse human KIR haplotypes. J Exp Med. 2001;193:135. [PMC free article: PMC2195888] [PubMed: 11136827]
- 103.
- Grendell R L , Hughes A L , Golos T G . Cloning of rhesus monkey killer-cell Ig-like receptors (KIRs) from early pregnancy decidua. Tissue Antigens. 2001;58:329. [PubMed: 11844144]
- 104.
- Mager D L , McQueen K L , Wee V , Freeman J D . Evolution of natural killer cell receptors: coexistence of functional Ly49 and KIR genes in baboons. Curr Biol. 2001;11:626. [PubMed: 11369209]
- 105.
- Hershberger K L , Shyam R , Miura A , Letvin N L . Diversity of the killer cell Ig-like receptors of rhesus monkeys. J Immunol. 2001;166:4380. [PubMed: 11254692]
- 106.
- McQueen K L , Wilhelm B T , Harden K D , Mager D L . Evolution of NK receptors: a single Ly49 and multiple KIR genes in the cow. Eur J Immunol. 2002;32:810. [PubMed: 11870625]
- 107.
- LaBonte M L , Hershberger K L , Korber B , Letvin N L . The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol Rev. 2001;183:25. [PubMed: 11782245]
- 108.
- McQueen K , Parham P . Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol. 2002;14:615. [PubMed: 12183162]
- 109.
- Anderson S K , Ortaldo J R , McVicar D W . The ever-expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79. [PubMed: 11513154]
- 110.
- Westgaard I H , Berg S F , Orstavik S , Fossum S , Dissen E . Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28:1839. [PubMed: 9645365]
- 111.
- Barten R , Trowsdale J . The human Ly-49L gene. Immunogenetics. 1999;49:731. [PubMed: 10369937]
- 112.
- Gumperz J E , Litwin V , Phillips J H , Lanier L L , Parham P . The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133. [PMC free article: PMC2191933] [PubMed: 7532677]
- 113.
- Martin A M , Freitas E M , Witt C S , Christiansen F T . The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268. [PubMed: 10803839]
- 114.
- Adams E J , Cooper S , Thomson G , Parham P . Common chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes. Immunogenetics. 2000;51:410. [PubMed: 10866107]
- 115.
- Moretta A , Vitale M , Bottino C , Orengo A M , Morelli L , Augugliaro R , Barbaresi M , Ciccone E , Moretta L . P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597. [PMC free article: PMC2191136] [PubMed: 8340759]
- 116.
- Melero I , Salmeron A , Balboa M A , Aramburu J , Lopez-Botet M . Tyrosine kinase-dependent activation of human NK cell functions upon stimulation through a 58-kDa surface antigen selectively expressed on discrete subsets of NK cells and T lymphocytes. J Immunol. 1994;152:1662. [PubMed: 8120376]
- 117.
- Snyder G A , Brooks A G , Sun P D . Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci U S A. 1999;96:3864. [PMC free article: PMC22386] [PubMed: 10097129]
- 118.
- Maenaka K , Juji T , Stuart D I , Jones E Y . Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure Fold Des. 1999;7:391. [PubMed: 10196125]
- 119.
- Yen J H , Moore B E , Nakajima T , Scholl D , Schaid D J , Weyand C M , Goronzy J J . Major histocompatibility complex class I-recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193:1159. [PMC free article: PMC2193323] [PubMed: 11369787]
- 120.
- Boyington J C , Brooks A G , Sun P D . Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol Rev. 2001;181:66. [PubMed: 11513153]
- 121.
- Katz G , Markel G , Mizrahi S , Arnon T I , Mandelboim O . Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260. [PubMed: 11390475]
- 122.
- Ponte M , Cantoni C , Biassoni R , Tradori-Cappai A , Bentivoglio G , Vitale C , Bertone S , Moretta A , Moretta L , Mingari M C . Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua- associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci U S A. 1999;96:5674. [PMC free article: PMC21919] [PubMed: 10318943]
- 123.
- Rajagopalan S , Long E O . A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093. [PMC free article: PMC2193010] [PubMed: 10190900]
- 124.
- Lopez-Botet M , Navarro F , Llano M . How do NK cells sense the expression of HLA-G class Ib molecules? Semin Cancer Biol. 1999;9:19. [PubMed: 10092547]
- 125.
- Faure M , Long E O . KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168:6208. [PubMed: 12055234]
- 126.
- Yusa S , Catina T L , Campbell K S . SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol. 2002;168:5047. [PubMed: 11994457]
- 127.
- Falk C S , Steinle A , Schendel D J . Expression of HLA-C molecules confers target cell resistance to some non-major histocompatibility complex-restricted T cells in a manner analogous to allospecific natural killer cells. J Exp Med. 1995;182:1005. [PMC free article: PMC2192283] [PubMed: 7561674]
- 128.
- Mingari M C , Ponte M , Cantoni C , Vitale C , Schiavetti F , Bertone S , Bellomo R , Cappai A T , Biassoni R . HLA-class I-specific inhibitory receptors in human cytolytic T lymphocytes: molecular characterization, distribution in lymphoid tissues and co-expression by individual T cells. Int Immunol. 1997;9:485. [PubMed: 9138008]
- 129.
- Gumperz J E , Valiante N M , Parham P , Lanier L L , Tyan D . Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med. 1996;183:1817. [PMC free article: PMC2192483] [PubMed: 8666938]
- 130.
- Raulet D H , Vance R E , McMahon C W . Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol. 2001;19:291. [PubMed: 11244039]
- 131.
- Shilling H G , Young N , Guethlein L A , Cheng N W , Gardiner C M , Tyan D , Parham P . Genetic control of human NK cell repertoire. J Immunol. 2002;169:239. [PubMed: 12077250]
- 132.
- Uhrberg M , Valiante N M , Young N T , Lanier L L , Phillips J H , Parham P . The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: clones sharing identical alpha beta TCR rearrangement express highly diverse killer cell Ig-like receptor patterns. J Immunol. 2001;166:3923. [PubMed: 11238637]
- 133.
- Husain Z , Alper C A , Yunis E J , Dubey D P . Complex expression of natural killer receptor genes in single natural killer cells. Immunology. 2002;106:373. [PMC free article: PMC1782738] [PubMed: 12100725]
- 134.
- Chan H W , Kurago Z B , Stewart C A , Wilson M J , Martin M P , Mace B E , Carrington M , Trowsdale J , Lutz C T . DNA Methylation Maintains Allele-specific KIR Gene Expression in Human Natural Killer Cells. J Exp Med. 2003;197:245. [PMC free article: PMC2193817] [PubMed: 12538663]
- 135.
- Pietra G , Semino C , Cagnoni F , Boni L , Cangemi G , Frumento G , Melioli G . Natural killer cells lyse autologous herpes simplex virus infected targets using cytolytic mechanisms distributed clonotypically. J Med Virol. 2000;62:354. [PubMed: 11055246]
- 136.
- Carr W H , Little A M , Mocarski E , Parham P . NK cell-mediated lysis of autologous HCMV-infected skin fibroblasts is highly variable among NK cell clones and polyclonal NK cell lines. Clin Immunol. 2002;105:126. [PubMed: 12482387]
- 137.
- Mingari M C , Ponte M , Vitale C , Bellomo R , Moretta L . Expression of HLA class I-specific inhibitory receptors in human cytolytic T lymphocytes: a regulated mechanism that controls T-cell activation and function. Hum Immunol. 2000;61:44. [PubMed: 10658977]
- 138.
- Young N T , Rust N A , Dallman M J , Cerundolo V , Morris P J , Welsh K I . Independent contributions of HLA epitopes and killer inhibitory receptor expression to the functional alloreactive specificity of natural killer cells. Hum Immunol. 1998;59:700. [PubMed: 9796738]
- 139.
- Frohn C , Schlenke P , Kirchner H . The repertoire of HLA-Cw-specific NK cell receptors CD158 a/b (EB6 and GL183) in individuals with different HLA phenotypes. Immunology. 1997;92:567. [PMC free article: PMC1364164] [PubMed: 9497500]
- 140.
- Zimmer J , Donato L , Hanau D , Cazenave J P , Tongio M M , Moretta A , de la Salle H . Activity and phenotype of natural killer cells in peptide transporter (TAP)-deficient patients (type I bare lymphocyte syndrome). J Exp Med. 1998;187:117. [PMC free article: PMC2199183] [PubMed: 9419217]
- 141.
- Long E O , Rajagopalan S . HLA class I recognition by killer cell Ig-like receptors. Semin Immunol. 2000;12:101. [PubMed: 10764618]
- 142.
- Sawicki M W , Dimasi N , Natarajan K , Wang J , Margulies D H , Mariuzza R A . Structural basis of MHC class I recognition by natural killer cell receptors. Immunol Rev. 2001;181:52. [PubMed: 11513152]
- 143.
- Biassoni R , Falco M , Cambiaggi A , Costa P , Verdiani S , Pende D , Conte R , Di Donato C , Parham P , Moretta L . Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by "group 2" or "group 1" NK clones. J Exp Med. 1995;182:605. [PMC free article: PMC2192139] [PubMed: 7629517]
- 144.
- Colonna M , Spies T , Strominger J L , Ciccone E , Moretta A , Moretta L , Pende D , Viale O . Alloantigen recognition by two human natural killer cell clones is associated with HLA-C or a closely linked gene. Proc Natl Acad Sci U S A. 1992;89:7983. [PMC free article: PMC49839] [PubMed: 1518825]
- 145.
- Winter C C , Long E O . A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026. [PubMed: 9126959]
- 146.
- Richardson J , Reyburn H T , Luque, Vales-Gomez M , Strominger J L . Definition of polymorphic residues on killer Ig-like receptor proteins which contribute to the HLA-C binding site. Eur J Immunol. 2000;30:1480. [PubMed: 10820396]
- 147.
- Cella M , Longo A , Ferrara G B , Strominger J L , Colonna M . NK3-specific natural killer cells are selectively inhibited by Bw4- positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235. [PMC free article: PMC2191670] [PubMed: 7931060]
- 148.
- Cantoni C , Verdiani S , Falco M , Pessino A , Cilli M , Conte R , Pende D , Ponte M , Mikaelsson M S , Moretta L , Biassoni R . p49, a putative HLA class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur J Immunol. 1998;28:1980. [PubMed: 9645380]
- 149.
- Khakoo S I , Geller R , Shin S , Jenkins J A , Parham P . The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J Exp Med. 2002;196:911. [PMC free article: PMC2194033] [PubMed: 12370253]
- 150.
- Rojo S , Wagtmann N , Long E O . Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568. [PubMed: 9045932]
- 151.
- Vales-Gomez M , Reyburn H T , Mandelboim M , Strominger J L . Kinetics of interaction of HLA-C ligands with natural killer cell inhibitory receptors. Immunity. 1998;9:337. [PubMed: 9768753]
- 152.
- Vyas Y M , Mehta K M , Morgan M , Maniar H , Butros L , Jung S , Burkhardt J K , Dupont B . Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I-regulated noncytolytic and cytolytic interactions. J Immunol. 2001;167:4358. [PubMed: 11591760]
- 153.
- Davis D M , Chiu, Fassett M , Cohen G B , Mandelboim O , Strominger J L . The human natural killer cell immune synapse. Proc Natl Acad Sci U S A. 1999;96:15062. [PMC free article: PMC24773] [PubMed: 10611338]
- 154.
- Carlin L M , Eleme K , McCann F E , Davis D M . Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses. J Exp Med. 2001;194:1507. [PMC free article: PMC2193674] [PubMed: 11714757]
- 155.
- Tabiasco J , Espinosa E , Hudrisier D , Joly E , Fournie J J , Vercellone A . Active trans-synaptic capture of membrane fragments by natural killer cells. Eur J Immunol. 2002;32:1502. [PubMed: 11981839]
- 156.
- Fassett M S , Davis D M , Valter M M , Cohen G B , Strominger J L . Signaling at the inhibitory natural killer cell immune synapse regulates lipid raft polarization but not class I MHC clustering. Proc Natl Acad Sci U S A. 2001;98:14547. [PMC free article: PMC64719] [PubMed: 11724921]
- 157.
- Watzl C , Long E O . Natural killer cell inhibitory receptors block actin cytoskeleton-dependent recruitment of 2B4 (CD244) to lipid rafts. J Exp Med. 2003;197:77. [PMC free article: PMC2193803] [PubMed: 12515815]
- 158.
- Moretta A , Sivori S , Vitale M , Pende D , Morelli L , Augugliaro R , Bottino C , Moretta L . Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875. [PMC free article: PMC2192157] [PubMed: 7650491]
- 159.
- Arase H , Mocarski E S , Campbell A E , Hill A B , Lanier L L . Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323. [PubMed: 11950999]
- 160.
- Brown M G , Dokun A O , Heusel J W , Smith H R , Beckman D L , Blattenberger E A , Dubbelde C E , Stone L R , Scalzo A A , Yokoyama W M . Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934. [PubMed: 11340207]
- 161.
- Lee S H , Girard S , Macina D , Busa M , Zafer A , Belouchi A , Gros P , Vidal S M . Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001;28:42. [PubMed: 11326273]
- 162.
- Daniels K A , Devora G , Lai W C , O'Donnell C L , Bennett M , Welsh R M . Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29. [PMC free article: PMC2193438] [PubMed: 11435470]
- 163.
- Smith H R , Heusel J W , Mehta K , Kim S , Dorner B G , Naidenko O V , Iizuka K , Furukawa H , Beckman D L , Pingel J T , Scalzo A A , Fremont D H , Yokoyama W M . Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826. [PMC free article: PMC124383] [PubMed: 12060703]
- 164.
- Vivier E , Biron C A . Immunology. A pathogen receptor on natural killer cells. Science. 2002;296:1248. [PubMed: 12016296]
- 165.
- Hanke T , Takizawa H , McMahon C W , Busch D H , Pamer E G , Miller J D , Altman J D , Liu Y , Cado D , Lemonnier F A , Bjorkman P J , Raulet D H . Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity. 1999;11:67. [PubMed: 10435580]
- 166.
- Mandelboim O , Reyburn H T , Vales-Gomez M , Pazmany L , Colonna M , Borsellino G , Strominger J L . Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913. [PMC free article: PMC2192787] [PubMed: 9064351]
- 167.
- Peruzzi M , Wagtmann N , Long E O . A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585. [PMC free article: PMC2192820] [PubMed: 8879234]
- 168.
- Rajagopalan S , Long E O . The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185:1523. [PMC free article: PMC2196286] [PubMed: 9126935]
- 169.
- Kim J , Chwae Y J , Kim M Y , Choi H , Park J H , Kim S J . Molecular basis of HLA-C recognition by p58 natural killer cell inhibitory receptors. J Immunol. 1997;159:3875. [PubMed: 9378975]
- 170.
- Winter C C , Gumperz J E , Parham P , Long E O , Wagtmann N . Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571. [PubMed: 9670929]
- 171.
- Maenaka K , Juji T , Nakayama T , Wyer J R , Gao G F , Maenaka T , Zaccai N R , Kikuchi A , Yabe T , Tokunaga K , Tadokoro K , Stuart D I , Jones E Y , van der Merwe P A . Killer cell immunoglobulin receptors and T cell receptors bind peptide- major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem. 1999;274:28329. [PubMed: 10497191]
- 172.
- Zappacosta F , Borrego F , Brooks A G , Parker K C , Coligan J E . Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc Natl Acad Sci U S A. 1997;94:6313. [PMC free article: PMC21046] [PubMed: 9177214]
- 173.
- Malnati M S , Peruzzi M , Parker K C , Biddison W E , Ciccone E , Moretta A , Long E O . Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016. [PubMed: 7863326]
- 174.
- Mandelboim O , Wilson S B , Vales-Gomez M , Reyburn H T , Strominger J L . Self and viral peptides can initiate lysis by autologous natural killer cells. Proc Natl Acad Sci U S A. 1997;94:4604. [PMC free article: PMC20770] [PubMed: 9114037]
- 175.
- Bruhns P , Marchetti P , Fridman W H , Vivier E , Daeron M . Differential roles of N- and C-terminal immunoreceptor tyrosine-based inhibition motifs during inhibition of cell activation by killer cell inhibitory receptors. J Immunol. 1999;162:3168. [PubMed: 10092767]
- 176.
- Fry A M , Lanier L L , Weiss A . Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J Exp Med. 1996;184:295. [PMC free article: PMC2192670] [PubMed: 8691146]
- 177.
- Burshtyn D N , Scharenberg A M , Wagtmann N , Rajagopalan S , Berrada K , Yi T , Kinet J P , Long E O . Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity. 1996;4:77. [PMC free article: PMC2423819] [PubMed: 8574854]
- 178.
- Leibson P J . Signal transduction during natural killer cell activation: inside the mind of a killer. Immunity. 1997;6:655. [PubMed: 9208838]
- 179.
- Brumbaugh K M , Binstadt B A , Billadeau D D , Schoon R A , Dick C J , Ten R M , Leibson P J . Functional role for Syk tyrosine kinase in natural killer cell-mediated natural cytotoxicity. J Exp Med. 1997;186:1965. [PMC free article: PMC2199178] [PubMed: 9396765]
- 180.
- Spaggiari G M , Contini P , Carosio R , Arvigo M , Ghio M , Oddone D , Dondero A , Zocchi M R , Puppo F , Indiveri F , Poggi A . Soluble HLA class I molecules induce natural killer cell apoptosis through the engagement of CD8: evidence for a negative regulation exerted by members of the inhibitory receptor superfamily. Blood. 2002;99:1706. [PubMed: 11861287]
- 181.
- Selvakumar A , Steffens U , Dupont B . NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens. 1996;48:285. [PubMed: 8946682]
- 182.
- Miller J S , McCullar V . Human natural killer cells with polyclonal lectin and immunoglobulinlike receptors develop from single hematopoietic stem cells with preferential expression of NKG2A and KIR2DL2/L3/S2. Blood. 2001;98:705. [PubMed: 11468170]
- 183.
- Young N T , Uhrberg M , Phillips J H , Lanier L L , Parham P . Differential expression of leukocyte receptor complex-encoded Ig-like receptors correlates with the transition from effector to memory CTL. J Immunol. 2001;166:3933. [PubMed: 11238638]
- 184.
- Santourlidis S , Trompeter H I , Weinhold S , Eisermann B , Meyer K L , Wernet P , Uhrberg M . Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253. [PubMed: 12370356]
- 185.
- Parham P . The rise and fall of great class I genes. Semin Immunol. 1994;6:373. [PubMed: 7654994]
- 186.
- Lanier L L , Chang C , Azuma M , Ruitenberg J J , Hemperly J J , Phillips J H . Molecular and functional analysis of human natural killer cell- associated neural cell adhesion molecule (N-CAM/CD56). J Immunol. 1991;146:4421. [PubMed: 1710251]
- 187.
- Jacobs R , Hintzen G , Kemper A , Beul K , Kempf S , Behrens G , Sykora K W , Schmidt R E . CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. 2001;31:3121. [PubMed: 11592089]
- 188.
- Phillips J H , Lanier L L . Dissection of the lymphokine-activated killer phenomenon. Relative contribution of peripheral blood natural killer cells and T lymphocytes to cytolysis. J Exp Med. 1986;164:814. [PMC free article: PMC2188384] [PubMed: 3489062]
- 189.
- Fehniger, T. A., M. A. Cooper, G. J. Nuovo, M. Cella, F. Facchetti, M. Colonna, and M. A. Caligiuri. 2002. CD56bright Natural Killer Cells are Present in Human Lymph Nodes and are Activated by T cell Derived IL-2: a Potential New Link between Adaptive and Innate Immunity. Blood. [PubMed: 12480696]
- 190.
- Long E O . Regulation of immune responses through inhibitory receptors. Annu Rev Immunol. 1999;17:875. [PubMed: 10358776]
- 191.
- Bakker A B , Phillips J H , Figdor C G , Lanier L L . Killer cell inhibitory receptors for MHC class I molecules regulate lysis of melanoma cells mediated by NK cells, gamma delta T cells, and antigen-specific CTL. J Immunol. 1998;160:5239. [PubMed: 9605119]
- 192.
- Snyder M R , Muegge L O , Offord C , O'Fallon W M , Bajzer Z , Weyand C M , Goronzy J J . Formation of the killer Ig-like receptor repertoire on CD4+CD28null T cells. J Immunol. 2002;168:3839. [PubMed: 11937537]
- 193.
- Young N T , Uhrberg M . KIR expression shapes cytotoxic repertoires: a developmental program of survival. Trends Immunol. 2002;23:71. [PubMed: 11929129]
- 194.
- Namekawa T , Snyder M R , Yen J H , Goehring B E , Leibson P J , Weyand C M , Goronzy J J . Killer cell activating receptors function as costimulatory molecules on CD4+CD28null T cells clonally expanded in rheumatoid arthritis. J Immunol. 2000;165:1138. [PubMed: 10878393]
- 195.
- Zhong F , McCombs C C , Olson J M , Elston R C , Stevens F M , McCarthy C F , Michalski J P . An autosomal screen for genes that predispose to celiac disease in the western counties of Ireland. Nat Genet. 1996;14:329. [PubMed: 8896565]
- 196.
- Moodie S J , Norman P J , King A L , Fraser J S , Curtis D , Ellis H J , Vaughan R W , Ciclitira P J . Analysis of candidate genes on chromosome 19 in coeliac disease: an association study of the KIR and LILR gene clusters. Eur J Immunogenet. 2002;29:287. [PubMed: 12121272]
- 197.
- McMaster M T , Librach C L , Zhou Y , Lim K H , Janatpour M J , DeMars R , Kovats S , Damsky C , Fisher S J . Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol. 1995;154:3771. [PubMed: 7706718]
- 198.
- Ruggeri L , Capanni M , Casucci M , Volpi, Tosti A , Perruccio K , Urbani E , Negrin R S , Martelli M F , Velardi A . Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333. [PubMed: 10381530]
- 199.
- Salter R D , Chan H W , Tadikamalla R , Lawlor D A . Domain organization and sequence relationship of killer cell inhibitory receptors. Immunol Rev. 1997;155:175. [PubMed: 9059893]
- 200.
- McCann F E , Suhling K , Carlin L M , Eleme K , Taner S B , Yanagi K , Vanherberghen B , French P M , Davis D M . Imaging immune surveillance by T cells and NK cells. Immunol Rev. 2002;189:179. [PubMed: 12445274]
- 201.
- Vyas Y M , Maniar H , Dupont B . Visualization of signaling pathways and cortical cytoskeleton in cytolytic and noncytolytic natural killer cell immune synapses. Immunol Rev. 2002;189:161. [PubMed: 12445273]
- 202.
- Lanier L L . NK cell receptors. Annu Rev Immunol. 1998;16:359. [PubMed: 9597134]
- 203.
- Biassoni R , Cantoni C , Pende D , Sivori S , Parolini S , Vitale M , Bottino C , Moretta A . Human natural killer cell receptors and co-receptors. Immunol Rev. 2001;181:203. [PubMed: 11513142]
- 204.
- Moretta A , Bottino C , Vitale M , Pende D , Cantoni C , Mingari M C , Biassoni R , Moretta L . Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197. [PubMed: 11244035]
- 205.
- McQueen K L , Parham P . Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol. 2002;14:615. [PubMed: 12183162]
- 206.
- Trowsdale J . Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363. [PubMed: 11567627]
- 207.
- Chang C C , Ciubotariu R , Manavalan J S , Yuan J , Colovai A I , Piazza F , Lederman S , Colonna M , Cortesini R , Dalla-Favera R , Suciu-Foca N . Tolerization of dendritic cells by T(S) cells: the crucial role of inhibitory receptors ILT3 and ILT4. Nat Immunol. 2002;3:237. [PubMed: 11875462]
- 208.
- Chapman T L , Heikema A P , West, Jr A P , Bjorkman P J . Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2). Immunity. 2000;13:727. [PubMed: 11114384]
- 209.
- Willcox B E , Thomas L M , Chapman T L , Heikema A P , West, Jr A P , Bjorkman P J . Crystal structure of LIR-2 (ILT4) at 1.8 A: differences from LIR-1 (ILT2) in regions implicated in the binding of the Human Cytomegalovirus class I MHC homolog UL18. BMC Struct Biol. 2002;2:6. [PMC free article: PMC130215] [PubMed: 12390682]
- 210.
- Biassoni R , Cantoni C , Falco M , Verdiani S , Bottino C , Vitale M , Conte R , Poggi A , Moretta A , Moretta L . The human leukocyte antigen (HLA)-C-specific "activatory" or "inhibitory" natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. 1996;183:645. [PMC free article: PMC2192451] [PubMed: 8627176]
- 211.
- Ferrini S , Cambiaggi A , Meazza R , Sforzini S , Marciano S , Mingari M C , Moretta L . T cell clones expressing the natural killer cell-related p58 receptor molecule display heterogeneity in phenotypic properties and p58 function. Eur J Immunol. 1994;24:2294. [PubMed: 7925558]
- 212.
- Moretta A , Bottino C , Pende D , Tripodi G , Tambussi G , Viale O , Orengo A , Barbaresi M , Merli A , Ciccone E . Identification of four subsets of human CD3-CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J Exp Med. 1990;172:1589. [PMC free article: PMC2188758] [PubMed: 2147946]
- 213.
- Bottino C , Sivori S , Vitale M , Cantoni C , Falco M , Pende D , Morelli L , Augugliaro R , Semenzato G , Biassoni R , Moretta L , Moretta A . A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur J Immunol. 1996;26:1816. [PubMed: 8765026]
- 214.
- Yawata M . et al. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol. 2003;22:463–482. [PubMed: 12803322]
- 215.
- Marsh S G . et al. Killer-cell Immunoglobulin-like Receptor (KIR) Nomenclature Report, 2002. Hum Immunol. 2003;64:648–654. [PubMed: 12770798]