By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Cooper GM. The Cell: A Molecular Approach. 2nd edition. Sunderland (MA): Sinauer Associates; 2000.

The Cell: A Molecular Approach. 2nd edition.
Show detailsThe nuclear envelope separates the contents of the nucleus from the cytoplasm and provides the structural framework of the nucleus. The nuclear membranes, acting as barriers that prevent the free passage of molecules between the nucleus and the cytoplasm, maintain the nucleus as a distinct biochemical compartment. The sole channels through the nuclear envelope are provided by the nuclear pore complexes, which allow the regulated exchange of molecules between the nucleus and cytoplasm. The selective traffic of proteins and RNAs through the nuclear pore complexes not only establishes the internal composition of the nucleus, but also plays a critical role in regulating eukaryotic gene expression.
Structure of the Nuclear Envelope
The nuclear envelope has a complex structure, consisting of two nuclear membranes, an underlying nuclear lamina, and nuclear pore complexes (Figure 8.1). The nucleus is surrounded by a system of two concentric membranes, called the inner and outer nuclear membranes. The outer nuclear membrane is continuous with the endoplasmic reticulum, so the space between the inner and outer nuclear membranes is directly connected with the lumen of the endoplasmic reticulum. In addition, the outer nuclear membrane is functionally similar to the membranes of the endoplasmic reticulum (see Chapter 9) and has ribosomes bound to its cytoplasmic surface. In contrast, the inner nuclear membrane carries unique proteins that are specific to the nucleus.

Figure 8.1
The nuclear envelope. (A) An electron micrograph of a nucleus. The inner and outer nuclear membranes are joined at nuclear pore complexes (arrows). (B) An electron micrograph illustrating the continuity of the outer nuclear membrane with the endoplasmic (more...)
The critical function of the nuclear membranes is to act as a barrier that separates the contents of the nucleus from the cytoplasm. Like other cell membranes, the nuclear membranes are phospholipid bilayers, which are permeable only to small nonpolar molecules (see Figure 2.49). Other molecules are unable to diffuse through the phospholipid bilayer. The inner and outer nuclear membranes are joined at nuclear pore complexes, the sole channels through which small polar molecules and macromolecules are able to travel through the nuclear envelope (Figure 8.2). As discussed in the next section, the nuclear pore complex is a complicated structure that is responsible for the selective traffic of proteins and RNAs between the nucleus and the cytoplasm.

Figure 8.2
Electron micrograph showing nuclear pores. Many nuclear pores (arrows) are visible in this freeze-fracture preparation of the nuclear envelope. (Photo Researchers, Inc.)
Underlying the inner nuclear membrane is the nuclear lamina, a fibrous meshwork that provides structural support to the nucleus (Figure 8.3). The nuclear lamina is composed of one or more related proteins called lamins. Most mammalian cells, for example, contain four different lamins, designated A, B1, B2, and C. All the lamins are 60- to 80-kilodalton (kd) fibrous proteins that are related to the intermediate filament proteins of the cytoskeleton (see Chapter 11). Like other intermediate filament proteins, the lamins associate with each other to form filaments (Figure 8.4). The first stage of this association is the interaction of two lamins to form a dimer in which the α-helical regions of two polypeptide chains are wound around each other in a structure called a coiled coil. These lamin dimers then associate with each other to form the filaments that make up the nuclear lamina. The association of lamins with the inner nuclear membrane is facilitated by the posttranslational addition of lipid—in particular, prenylation of C-terminal cysteine residues (see Figure 7.30). In addition, the lamins bind to inner nuclear membrane proteins, which may help organize the lamin filaments into a meshwork and mediate their attachment to the membrane.

Figure 8.3
Electron micrograph of the nuclear lamina. The lamina is a meshwork of filaments underlying the inner nuclear membrane. (From U. Aebi, J. Cohn, L. Buhle and L. Gerace, 1986. Nature 323: 560.)
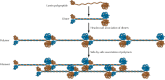
Figure 8.4
Model of lamin assembly. The lamin polypeptides form dimers in which the central α-helical regions of two polypeptide chains are wound around each other. Further assembly may involve the head-to-tail association of dimers to form linear polymers (more...)
In addition to providing structural support to the nucleus, the nuclear lamina is thought to serve as a site of chromatin attachment. Chromatin within the nucleus is organized into large loops of DNA, some of which appear to be bound to the nuclear envelope. The lamins bind chromatin and may help mediate this interaction.
The Nuclear Pore Complex
The nuclear pore complexes are the only channels through which small polar molecules, ions, and macromolecules (proteins and RNAs) are able to travel between the nucleus and the cytoplasm. The nuclear pore complex is an extremely large structure with a diameter of about 120 nm and an estimated molecular mass of approximately 125 million daltons—about 30 times the size of a ribosome. In vertebrates, the nuclear pore complex is composed of 50 to 100 different proteins. By controlling the traffic of molecules between the nucleus and cytoplasm, the nuclear pore complex plays a fundamental role in the physiology of all eukaryotic cells. RNAs that are synthesized in the nucleus must be efficiently exported to the cytoplasm, where they function in protein synthesis. Conversely, proteins required for nuclear functions (e.g., transcription factors) must be transported into the nucleus from their sites of synthesis in the cytoplasm. In addition, many proteins shuttle continuously between the nucleus and the cytoplasm. The regulated traffic of proteins and RNAs through the nuclear pore complex thus determines the composition of the nucleus and plays a key role in gene expression.
Depending on their size, molecules can travel through the nuclear pore complex by one of two different mechanisms (Figure 8.5). Small molecules and some proteins with molecular mass less than approximately 50 kd pass freely across the nuclear envelope in either direction: cytoplasm to nucleus or nucleus to cytoplasm. These molecules diffuse passively through open aqueous channels, estimated to have diameters of approximately 9 nm, in the nuclear pore complex. Most proteins and RNAs, however, are unable to pass through these open channels. Instead, these macromolecules pass through the nuclear pore complex by an active process in which appropriate proteins and RNAs are recognized and selectively transported in only one direction (nucleus to cytoplasm or cytoplasm to nucleus). The traffic of these molecules occurs through regulated channels in the nuclear pore complex that, in response to appropriate signals, can open to a diameter of more than 25 nm—a size sufficient to accommodate large ribonucleoprotein complexes, such as ribosomal subunits. It is through these regulated channels that nuclear proteins are selectively imported from the cytoplasm to the nucleus while RNAs are exported from the nucleus to the cytoplasm.
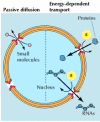
Figure 8.5
Molecular traffic through nuclear pore complexes. Small molecules are able to pass rapidly through open channels in the nuclear pore complex by passive diffusion. In contrast, macromolecules are transported by a selective, energy-dependent mechanism that (more...)
Visualization of nuclear pore complexes by electron microscopy reveals a structure with eightfold symmetry organized around a large central channel (Figure 8.6), which is the route through which proteins and RNAs cross the nuclear envelope. Detailed structural studies, including computer-based image analysis, have led to the development of three-dimensional models of the nuclear pore complex (Figure 8.7). These studies indicate that the nuclear pore complex consists of an assembly of eight spokes arranged around a central channel. The spokes are connected to rings at the nuclear and cytoplasmic surfaces, and the spoke-ring assembly is anchored within the nuclear envelope at sites of fusion between the inner and outer nuclear membranes. Protein filaments extend from both the cytoplasmic and nuclear rings, forming a distinct basketlike structure on the nuclear side. The central channel is approximately 40 nm in diameter, which is wide enough to accommodate the largest particles able to cross the nuclear envelope. It contains a structure called the central transporter, through which the active transport of macromolecules is thought to occur.

Figure 8.6
Electron micrograph of nuclear pore complexes. In this face-on view, isolated nuclear pore complexes appear to consist of eight structural subunits surrounding a central channel. (Courtesy of Dr. Ron Milligan, Scripps Research Institute.)
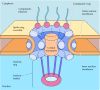
Figure 8.7
Model of the nuclear pore complex. The complex consists of an assembly of eight spokes attached to rings on the cytoplasmic and nuclear sides of the nuclear envelope. The spoke-ring assembly surrounds a central channel containing the central transporter. (more...)
Selective Transport of Proteins to and from the Nucleus
The basis for selective traffic across the nuclear envelope is best understood for proteins that are imported from the cytoplasm to the nucleus. Such proteins are responsible for all aspects of genome structure and function; they include histones, DNA polymerases, RNA polymerases, transcription factors, splicing factors, and many others. These proteins are targeted to the nucleus by specific amino acid sequences, called nuclear localization signals, that direct their transport through the nuclear pore complex.
The first nuclear localization signal to be mapped in detail was characterized by Alan Smith and colleagues in 1984. These investigators studied simian virus 40 (SV40) T antigen, a virus-encoded protein that initiates viral DNA replication in infected cells (see Chapter 5). As expected for a replication protein, T antigen normally is localized to the nucleus. The signal responsible for its nuclear localization was first identified by the finding that mutation of a single lysine residue prevents nuclear import, resulting instead in the accumulation of T antigen in the cytoplasm. Subsequent studies defined the T antigen nuclear localization signal as the seven-amino-acid sequence Pro-Lys-Lys-Lys-Arg-Lys-Val. Not only was this sequence necessary for the nuclear transport of T antigen, but its addition to other, normally cytoplasmic, proteins was also sufficient to direct their accumulation in the nucleus.
Nuclear localization signals have since been identified in many other proteins. Most of these sequences, like that of T antigen, are short stretches rich in basic amino acid residues (lysine and arginine). In many cases, however, the amino acids that form the nuclear localization signal are close together but not immediately adjacent to each other. For example, the nuclear localization signal of nucleoplasmin (a protein involved in chromatin assembly) consists of two parts: a Lys-Arg pair followed by four lysines located ten amino acids farther downstream (Figure 8.8). Both the Lys-Arg and Lys-Lys-Lys-Lys sequences are required for nuclear targeting, but the ten amino acids between these sequences can be mutated without affecting nuclear localization. Because this nuclear localization sequence is composed of two separated elements, it is referred to as bipartite. Similar bipartite motifs appear to function as the localization signals of many nuclear proteins; thus they may be more common than the simpler nuclear localization signal of T antigen. In addition, some proteins, such as ribosomal proteins, contain distinct nuclear localization signals which are unrelated to the basic amino acid-rich nuclear localization signals of either nucleoplasmin or T antigen.
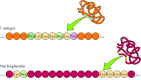
Figure 8.8
Nuclear localization signals. The T antigen nuclear localization signal is a single stretch of amino acids. In contrast, the nuclear localization signal of nucleoplasmin is bipartite, consisting of a Lys-Arg sequence, followed by a Lys-Lys-Lys-Lys sequence (more...)
Protein import through the nuclear pore complex can be operationally divided into two steps, distinguished by whether they require energy (Figure 8.9). In the first step, which does not require energy, proteins that contain nuclear localization signals bind to the nuclear pore complex but do not pass through the pore. In this initial step, nuclear localization signals are recognized by a cytosolic receptor protein, and the receptor-substrate complex binds to the nuclear pore. The prototype receptor, called importin, consists of two subunits. One subunit (importin α) binds to the basic amino acid-rich nuclear localization signals of proteins such as T antigen and nucleoplasmin. The second subunit (importin β) binds to the cytoplasmic filaments of the nuclear pore complex, bringing the target protein to the nuclear pore. Other types of nuclear localization signals, such as those of ribosomal proteins, are recognized by distinct receptors which are related to importin β and function similarly to importin β during the transport of their target proteins into the nucleus.
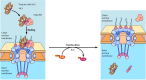
Figure 8.9
Protein import through the nuclear pore complex. Proteins are transported through the nuclear pore complex in two steps. In the example shown, a protein with a classical basic amino acid-rich nuclear localization sequence (NLS) is recognized by importin (more...)
The second step in nuclear import, translocation through the nuclear pore complex, is an energy-dependent process that requires GTP hydrolysis. A key player in the translocation process is a small GTP-binding protein called Ran, which is related to the Ras proteins (Figure 8.10). The conformation and activity of Ran is regulated by GTP binding and hydrolysis, like Ras or several of the translation factors involved in protein synthesis (see Figure 7.12). Enzymes that stimulate GTP binding to Ran are localized to the nuclear side of the nuclear envelope whereas enzymes that stimulate GTP hydrolysis are localized to the cytoplasmic side. Consequently, there is a gradient of Ran/GTP across the nuclear envelope, with a high concentration of Ran/GTP in the nucleus and a high concentration of Ran/GDP in the cytoplasm. This gradient of Ran/GTP is thought to determine the directionality of nuclear transport, and GTP hydrolysis by Ran appears to account for most (if not all) of the energy required for nuclear import. Importin β forms a complex with importin α and its associated target protein on the cytoplasmic side of the nuclear pore complex, in the presence of a high concentration of Ran/ GDP. This complex is then transported through the nuclear pore to the nucleus, where a high concentration of Ran/GTP is present. At the nuclear side of the pore, Ran/GTP binds to importin β, displacing importin α and the target protein. As a result, the target protein is released within the nucleus. The Ran/GTP-importin β complex is then exported to the cytosol, where the bound GTP is hydrolyzed to GDP, releasing importin β to participate in another cycle of nuclear import.
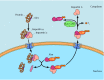
Figure 8.10
Role of the Ran protein in nuclear import. Transport through the nuclear pore complex is driven by a gradient of Ran/GTP, with a high concentration of Ran/GDP in the cytoplasm and a high concentration of Ran/GTP in the nucleus. Complexes form between (more...)
Some proteins remain within the nucleus following their import from the cytoplasm, but many others shuttle back and forth between the nucleus and the cytoplasm. Some of these proteins act as carriers in the transport of other molecules, such as RNAs; others coordinate nuclear and cytoplasmic functions (e.g., by regulating the activities of transcription factors). Proteins are targeted for export from the nucleus by specific amino acid sequences, called nuclear export signals. Like nuclear localization signals, nuclear export signals are recognized by receptors within the nucleus that direct protein transport through the nuclear pore complex to the cytoplasm. Interestingly, the nuclear export receptors (called exportins) are related to importin β. Like importin β, the exportins bind to Ran, which is required for nuclear export as well as for nuclear import (Figure 8.11). Strikingly, however, Ran/GTP promotes the formation of stable complexes between exportins and their target proteins, whereas it dissociates the complexes between importins and their targets. This effect of Ran/GTP binding on exportins dictates the movement of proteins containing nuclear export signals from the nucleus to the cytoplasm. Thus, exportins form stable complexes with their target proteins in association with Ran/GTP within the nucleus. Following transport to the cytosolic side of the nuclear envelope, GTP hydrolysis leads to dissociation of the target protein, which is released into the cytoplasm.

Figure 8.11
Nuclear export. Complexes between target proteins bearing nuclear export signals (NES), exportins, and Ran/ GTP form in the nucleus. Following transport through the nuclear pore complex, Ran GAP stimulates the hydrolysis of bound GTP, leading to formation (more...)
Regulation of Nuclear Protein Import
An intriguing aspect of the transport of proteins into the nucleus is that it is another level at which the activities of nuclear proteins can be controlled. Transcription factors, for example, are functional only when they are present in the nucleus, so regulation of their import to the nucleus is a novel means of controlling gene expression. As will be discussed in Chapter 13, the regulated nuclear import of both transcription factors and protein kinases plays an important role in controlling the behavior of cells in response to changes in the environment, because it provides a mechanism by which signals received at the cell surface can be transmitted to the nucleus.
In one mechanism of regulation, transcription factors (or other proteins) associate with cytoplasmic proteins that mask their nuclear localization signals; because their signals are no longer recognizable, these proteins remain in the cytoplasm. A good example is provided by the transcription factor NF-κB, which activates transcription of immunoglobulin-κ light chains in B lymphocytes (Figure 8.12). In unstimulated cells, NF-κB is found as an inactive complex with an inhibitory protein (IκB) in the cytoplasm. Binding to IκB appears to mask the NF-κB nuclear localization signal, thus preventing NF-κB from being transported into the nucleus. In stimulated cells, IκB is phosphorylated and degraded by ubiquitin-mediated proteolysis, allowing NF-κB to enter the nucleus and activate transcription of its target genes.
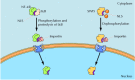
Figure 8.12
Regulation of nuclear import of transcription factors. The transcription factor NF-κB is maintained as an inactive complex with IκB, which masks its nuclear localization sequence (NLS), in the cytoplasm. In response to appropriate extracellular (more...)
The nuclear import of other transcription factors is regulated directly by their phosphorylation, rather than by association with inhibitory proteins. For example, the yeast transcription factor SWI5 is imported into the nucleus only at a specific stage of the cell cycle (see Figure 8.12). Otherwise, SWI5 is retained in the cytoplasm as a result of phosphorylation at serine residues adjacent to its nuclear localization signal, preventing nuclear import. Regulated dephosphorylation of these sites activates SWI5 at the appropriate stage of the cell cycle by permitting its translocation to the nucleus.
Transport of RNAs
Whereas many proteins are selectively transported from the cytoplasm into the nucleus, most RNAs are exported from the nucleus to the cytoplasm. Since proteins are synthesized in the cytoplasm, the export of mRNAs, rRNAs, and tRNAs is a critical step in gene expression in eukaryotic cells. Like protein import, the export of RNAs through nuclear pore complexes is an active, energy-dependent process that requires the Ran GTP-binding protein.
RNAs are transported across the nuclear envelope as RNA-protein complexes, which in some cases are large enough to visualize by electron microscopy (Figure 8.13). The substrates for transport are ribonucleoprotein complexes rather than naked RNAs, and RNAs are targeted for transport from the nucleus by nuclear export signals on the proteins bound to them. These proteins are recognized by exportins and transported from the nucleus to the cytoplasm as described earlier (see Figure 8.11). Pre-mRNAs and mRNAs are associated with a set of at least 20 proteins (forming heterogeneous nuclear ribonucleoproteins, or hnRNPs) throughout their processing in the nucleus and eventual transport to the cytoplasm. At least two of these hnRNP proteins contain nuclear export signals and are thought to function as carriers of mRNAs during their export to the cytoplasm. As discussed in a later section of this chapter, ribosomal RNAs are assembled with ribosomal proteins in the nucleolus, and intact ribosomal subunits are then transported to the cytoplasm. Their export from the nucleus appears to be mediated by nuclear export signals present on ribosomal proteins. For tRNAs, the specific proteins that mediate nuclear export remain to be identified.

Figure 8.13
Transport of a ribonucleoprotein complex. Insect salivary gland cells produce large ribonucleoprotein complexes (RNPs), which contain 35 to 40 kb of RNA and have a total mass of approximately 30 million daltons. This series of electron micrographs shows (more...)
In contrast to mRNAs, tRNAs, and rRNAs, which function in the cytoplasm, the snRNAs function within the nucleus as components of the RNA processing machinery. Perhaps surprisingly, these RNAs are initially transported from the nucleus to the cytoplasm, where they associate with proteins to form functional snRNPs and then return to the nucleus (Figure 8.14). Proteins that bind to the 5′ caps of snRNAs appear to be involved in the export of the snRNAs to the cytoplasm, whereas sequences present on the snRNP proteins are responsible for the transport of snRNPs from the cytoplasm to the nucleus.
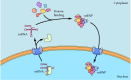
Figure 8.14
Transport of snRNAs between nucleus and cytoplasm. Small nuclear RNAs are initially exported from the nucleus to the cytoplasm, where they associate with proteins to form snRNPs. The assembled snRNPs are then transported back into the nucleus.
Box
Key Experiment: Identification of Nuclear Localization Signals.
- The Nuclear Envelope and Traffic between the Nucleus and Cytoplasm - The CellThe Nuclear Envelope and Traffic between the Nucleus and Cytoplasm - The Cell
- ARV1_gp23 [Acidianus rod-shaped virus 1]ARV1_gp23 [Acidianus rod-shaped virus 1]Gene ID:5729550Gene
Your browsing activity is empty.
Activity recording is turned off.
See more...