query structure | similar structures | filters ( complete biounit matches, complete biounit matches,  partial biounit matches, taxonomy) | folder tabs to select 3D alignment type (all matching molecules superposed, invariant substructure superposed) | ranked list of hits (sort options, view details for a similar structure, search within results) | "original VAST" button | null result: "no matched structure found" | "new search" button partial biounit matches, taxonomy) | folder tabs to select 3D alignment type (all matching molecules superposed, invariant substructure superposed) | ranked list of hits (sort options, view details for a similar structure, search within results) | "original VAST" button | null result: "no matched structure found" | "new search" button
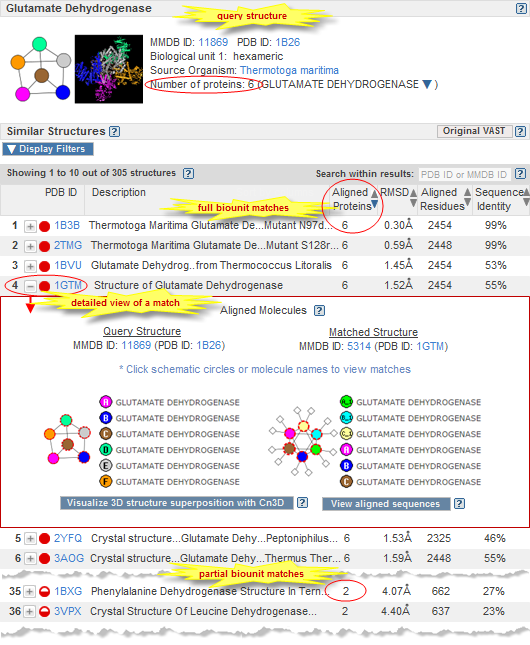
The VAST+ search results page displays summary information about your query structure, followed by a list of similar structures that are ranked based on their degree of similarity to the query structure's macromolecular complex (biological unit) (illustrated examples).
By default, all similar structures are listed, starting with  complete matches to the query structure's biological unit, followed by complete matches to the query structure's biological unit, followed by  partial matches, and ending with matches to individual protein molecules.
If desired, you can activate a subset of filters in order to display only the subset of similar structures that meet the specified criteria. It is also possible to change the sort order of the similar structures, and to search within results to see if/where a specific structure falls within the search results.
Click on the plus button partial matches, and ending with matches to individual protein molecules.
If desired, you can activate a subset of filters in order to display only the subset of similar structures that meet the specified criteria. It is also possible to change the sort order of the similar structures, and to search within results to see if/where a specific structure falls within the search results.
Click on the plus button  beside any structure to see a detailed view comparing the query structure to a matched structure. (If you prefer to view your search results in the original style VAST display, which lists structures that have similarities to individual protein molecules, or similarities to individual 3D domains, in your query structure, click the "Original VAST" button.) beside any structure to see a detailed view comparing the query structure to a matched structure. (If you prefer to view your search results in the original style VAST display, which lists structures that have similarities to individual protein molecules, or similarities to individual 3D domains, in your query structure, click the "Original VAST" button.)
A VAST+ search results page includes the following components:
 
The top section of a VAST+ results display provides the following summary information about the query structure, including:
interactions schematic |
molecular graphic |
MMDB ID, PDB ID |
biological unit |
source organism |
number of proteins |
number of nucleotides |
number of chemicals
Interactions schematic
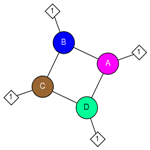 |
The interactions schematic shows the molecular components of the query structure's biological unit and the interactions among them.
The molecular components of the biological unit can include the following:
| Proteins, if present, are shown as circles: |
 etc. etc. |
|
 |
| Nucleotide sequences (DNA, RNA), if present, are shown as squares: |
 etc. etc. |
|
 |
| Chemicals, if present, are shown as diamonds: |
 etc. etc. |
|
 |
If any protein or nucleotide molecules in the structure were generated by applying transformations from crystallographic symmetry, their labels are shown as alphanumeric combinations (for example,  or or  ), indicating the source molecule from which they were generated and the copy number. Chemicals that interact only with such molecules were also generated by applying transformations from crystallographic symmetry. ), indicating the source molecule from which they were generated and the copy number. Chemicals that interact only with such molecules were also generated by applying transformations from crystallographic symmetry. |
The protein and nucleotide icons are scaled to show the relative sizes of those molecular components, so they are roughly comparable to each other based on molecular weight. All chemical icons are the same size.
Interactions among components are shown as lines, and an interaction is displayed only if there are at least 5 contacts at a distance of 4 Å or less between the heavy atoms of the molecules. (There is no meaning to the length of the lines in the interaction schematic. After the interactions are drawn, the diagram is flattened out to fit into the square, lengthening or shortening lines as needed.)
Because of the latter thresholds, ions that are part of the biological unit may be missing from the interaction diagram, but they will be listed in the table of molecular components and interactions on the query structure's MMDB summary page. Interactions for short peptides, or for molecule types other than protein, DNA/RNA, and chemical, are not calculated. Molecules, such as crystallization agents, etc., that are not part of the biologically active molecule are absent from both the interaction schematic and the molecular components list.
Mouse over any node in the schematic to view the molecule name.
|
Molecular graphic
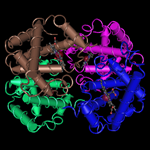 |
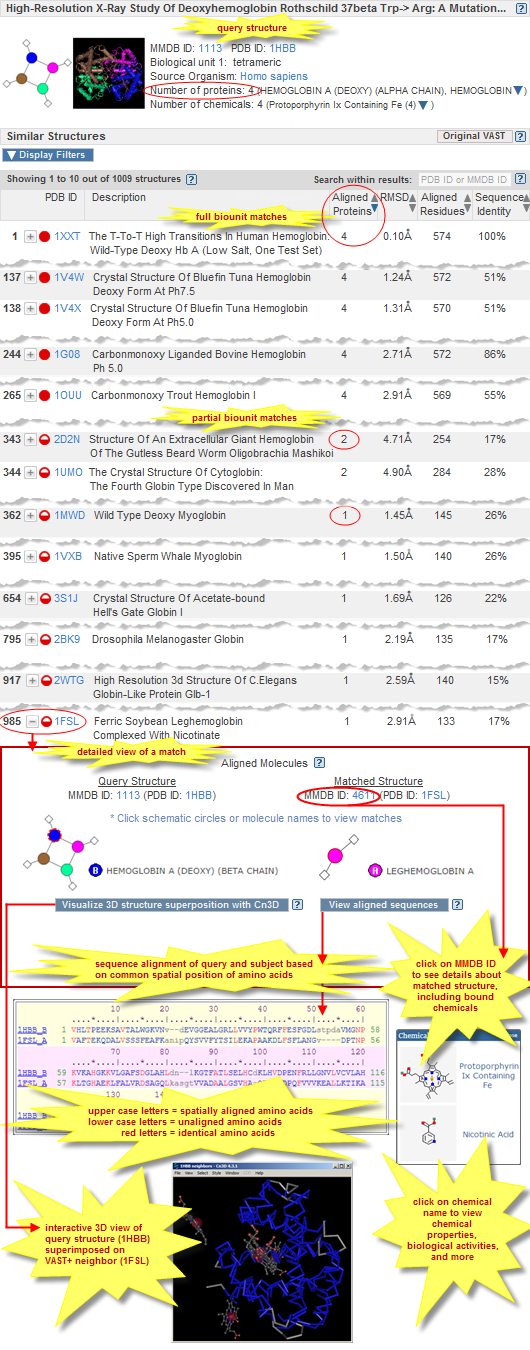
The 3D molecular graphic shows a single static snapshot of the query structure's 3D shape, generated by the Cn3D program.
In general, it shows the default biological unit of the structure.
To view the query's 3D structure interactively, click on its MMDB ID. That will open the corresponding structure summary page in MMDB, where you can choose to "View structure."
To view a 3D superposition of the query structure and any one of the VAST+ hits, Click on the plus button  beside a hit to open the detailed view comparing the query structure to a matched structure, then click the "Visualize 3D structure superposition with Cn3D" button at the bottom of that view. beside a hit to open the detailed view comparing the query structure to a matched structure, then click the "Visualize 3D structure superposition with Cn3D" button at the bottom of that view.
In either case, the interactive 3D view will open in iCn3D, NCBI's WebGL-based viewer.
There is no need to install a separate application in order to use iCn3D; you just need to use a web browser that supports WebGL. If your browser doesn't support WebGL, you might need to modify the settings in the browser to enable WebGL, or update your web browser to a newer version that supports WebGL. (See the WebGL site for more information about compatibility with various web browsers.)
|
|
MMDB ID and PDB ID |
MMDB ID is the unique identifier of the structure record in the Molecular Modeling Database (MMDB). It is a string of digits (e.g., 50885 for sheep prostaglandin H2 synthase) that are assigned consecutively to each structure record processed by NCBI.
PDB ID is the accession of number of the Protein Data Bank (PDB) record from which the corresponding MMDB record was derived. It is generally an alphanumeric combination (e.g., 1PTH, which served as the source record for MMDB ID 50885).
|
|
Biological Unit |
The biological unit ("biounit") of a structure is the biochemically active form of a biomolecule. It can range from a monomer (single protein molecule) to an oligomer of 100+ protein molecules, and is sometimes also referred to as the "biological assembly" or "macromolecular complex," reflecting the molecule's quaternary structure.
The query structure's biological unit, if available, is included in the summary information at the top of a VAST+ results page. (To see the biological unit of any individual VAST neighbor, and other details about the match, click on the plus button  beside the structure of interest.) beside the structure of interest.)
The data processing section of the MMDB help document provides additional information about biological units, incuding the procedures used to identify biological units.
If the query structure does not have a biological unit defined in its source PDB file, it is only available as an asymmetric unit (i.e., raw structure data). In that case, the structure is not processed by VAST+, since VAST+ focuses on finding similarities among biological units. However, such structures are processed by the Original VAST algorithm, which may have found structures that are geometrically similar to individual proteins, or individual 3D domains, in your query. (Read more about the occasional cirsumstances in which a query structure might generate a null result in VAST+).
|
|
Source Organism |
The source organism(s) of the protein and/or nucleotide molecules in the structure record. If a structure record contains protein or nucleotide sequences from more than one organism (e.g., human AND HIV1), each source organism is listed and links to the corresponding taxonomic information in the NCBI Taxonomy database, including the organism's Taxonomy ID (TaxID) and lineage. |
|
Number of proteins |
The total number of protein molecules in the biological unit of the query structure.
For brevity, only the first protein name is displayed by default.
Click on the down arrow  to view the complete list of proteins in the biological unit. to view the complete list of proteins in the biological unit.
The MMDB help document provides additional information about the molecular components of a structure, incuding its proteins. |
|
Number of nucleotides |
The total number of nucleotide molecules (DNA and/or RNA) in the biological unit of the query structure.
For brevity, only the first nucleotide molecule's name is displayed by default.
Click on the down arrow  to view the complete list of nucleotide molecules in the biological unit. to view the complete list of nucleotide molecules in the biological unit.
The MMDB help document provides additional information about the molecular components of a structure, incuding its nucleotide sequences.
|
|
Number of chemicals |
The total number of chemicals in the biological unit of the query structure.
For brevity, only the first chemical name is displayed by default.
Click on the down arrow  to view the complete list of chemicals in the biological unit. to view the complete list of chemicals in the biological unit.
The MMDB help document provides additional information about the molecular components of a structure, incuding its chemicals.
|
 
The VAST+ search results appear immediately beneath the query structure summary information, and include the following features and functions:
 
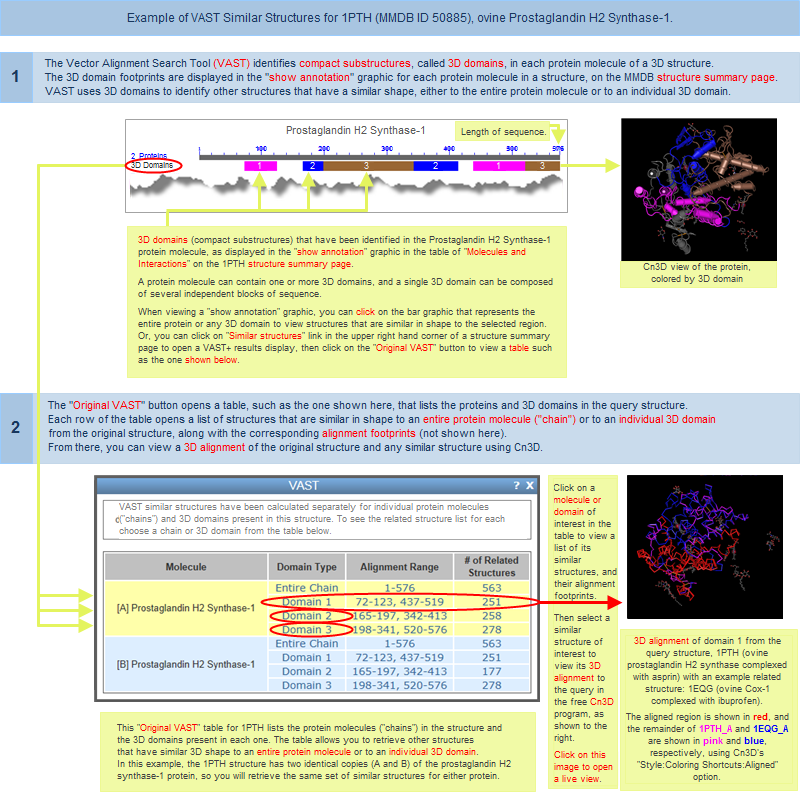
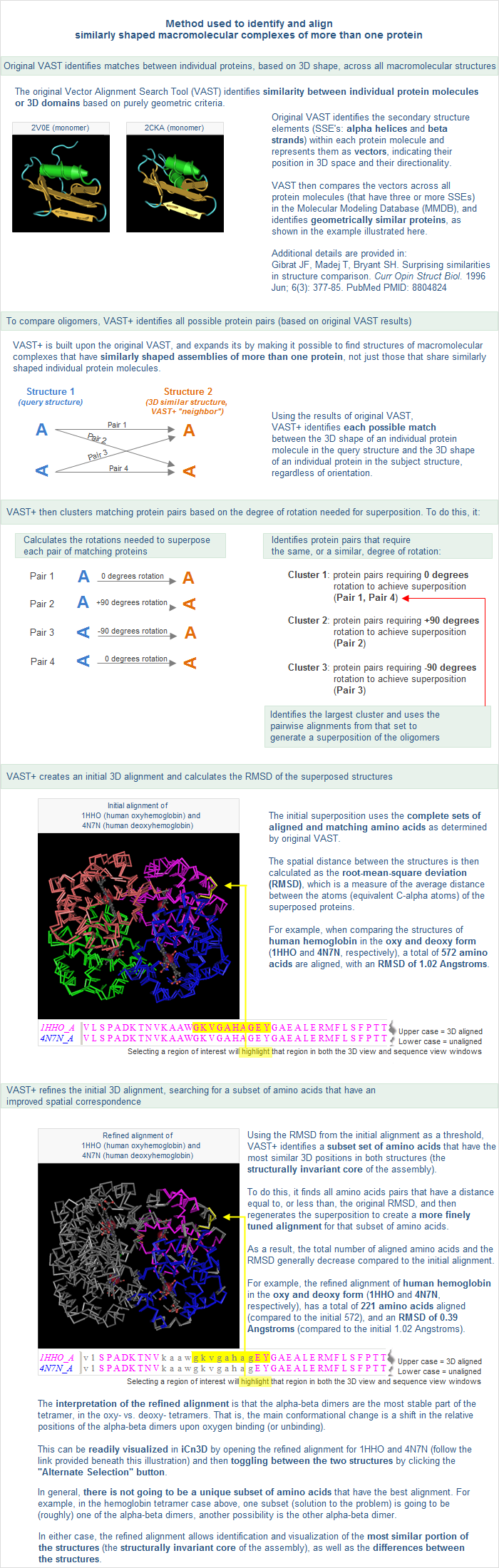
The VAST algorithm calculates two different types of 3D alignments between a pair of similar structures: an initial alignment that shows all matching molecules superposed, and a refined alignment that shows the invariant substructure superposed.
Each of the following folder tabs on the VAST+ output page includes the complete set of search results, listing all of structures that VAST+ found to be similar to your query structure.
However, a given structure might have a different rank in the first tab versus the second tab, because the statistics generated for that match (e.g., RMSD and number of aligned residues) generally differ between the two alignment types, and those statistics are used to determine the sort order of the search results.
- All matching molecules superposed (initial alignment)
- When you click on the plus button
 beside any structure to see a detailed view/graphical display comparing the query structure to the matched structure, the "visualize 3D structure superposition" button will open the initial 3D alignment, with all matching molecules superposed. The VAST algorithm section of this document provides more details about the initial alignment as well as an example. beside any structure to see a detailed view/graphical display comparing the query structure to the matched structure, the "visualize 3D structure superposition" button will open the initial 3D alignment, with all matching molecules superposed. The VAST algorithm section of this document provides more details about the initial alignment as well as an example.
- Invariant substructure superposed (refined alignment)
- When you click on the plus button
 beside any structure to see a detailed view/graphical display comparing the query structure to the matched structure, the "visualize 3D structure superposition" button will open the refined 3D alignment, with the invariant substructure superposed. The VAST algorithm section of this document provides more details about the refined alignment as well as an example. beside any structure to see a detailed view/graphical display comparing the query structure to the matched structure, the "visualize 3D structure superposition" button will open the refined 3D alignment, with the invariant substructure superposed. The VAST algorithm section of this document provides more details about the refined alignment as well as an example.
|
 
An option to Display Filters appears above the ranked list of similar structures. This option generates bar graphs that: (1) provide aggregate information about the structures; (2) categorize the retrieved structures by their degree of similarity to the query structure and their taxonomic distribution; and (3) enable you to subset the results.
By default, all similar structures are listed in the results display. Activate the check boxes beside one or more bar graphs to view only the subset of similar structures that meet the criteria you selected. (Note: Selecting all of the check boxes is the same as selecting none (default), and will display the complete set of results. Selecting a subset of the checkboxes will display the subset of results that meet the desired criteria.)
- Filter by number of matching molecules
 
-
 Complete match to biological unit of query structure -- Each of the biopolymer molecules in the biological unit of the query structure have a match (i.e., are similar in 3D shape) to a biopolymer molecule in the subject. The illustrated examples of VAST+ results include complete matches, which are indicated by solid red circles. Complete match to biological unit of query structure -- Each of the biopolymer molecules in the biological unit of the query structure have a match (i.e., are similar in 3D shape) to a biopolymer molecule in the subject. The illustrated examples of VAST+ results include complete matches, which are indicated by solid red circles.
 Partial match to biological unit of query structure -- A subset of the biopolymer molecules, or an individual biopolymer molecule, from the biological unit of the query structure have a match (i.e., are similar in 3D shape) to a biopolymer molecule in the subject. The illustrated examples of VAST+ results include partial matches, which are indicated by half-filled red circles. Partial match to biological unit of query structure -- A subset of the biopolymer molecules, or an individual biopolymer molecule, from the biological unit of the query structure have a match (i.e., are similar in 3D shape) to a biopolymer molecule in the subject. The illustrated examples of VAST+ results include partial matches, which are indicated by half-filled red circles.
- Additional notes about matching molecules:
- Biopolymers refer to protein, DNA, and RNA molecules.
- At this time, VAST (and VAST+) operates only on protein molecules. Some biological units might contain other types of biopolymers as well, such as DNA or RNA molecules. VAST and VAST+ do not currently compute similarities between the 3D shapes of DNA molecules, or similarities between the 3D shapes of RNA molecules.
Example: 1TUP, "Tumor Suppressor P53 Complexed With DNA," has a pentameric biological unit composed of three proteins and two nucleotide sequences. VAST will find other structures that have similarly shaped proteins, but will not take the DNA molecules into consideration. Therefore, there will be no "complete match" structures in the VAST+ search results for 1TUP, as it is not currently possible to find a match for all five molecules in the biological unit. The top scoring hits, which match all three protein molecules in 1TUP, are nevertheless easy to find as they are displayed at the top of the "partial match" results, based on the default sort order.
- Mimimum size of protein: VAST does not act upon protein molecules that have fewer than three secondary structure elements (SSE's: alpha helices and/or beta sheets) because they can generate spurious hits. Such short proteins are therefore not considered in VAST+ calculations or results. For example, if a pentameric structure contains four large proteins, plus one small protein that doesn't meet the VAST size requirements, VAST will only operate on the four large proteins from the biological unit. Therefore, there will be no "complete match" structures in the VAST+ search results, and the top hits will be, at the most, structures that match all four long proteins.
Example: 2YPL, "Structural Features Underlying T-cell Receptor Sensitivity to Concealed MHC Class I Micropolymorphisms," contains four human proteins, plus one very small HIV protein. None of the structures in the VAST+ search results for 2YPL are listed as a complete hit, because the fifth protein in the 2YPL's biological unit does not meet the minimum size requirements for VAST. So the best possible match that VAST (and VAST+) can find is to 4 out of 5 proteins in the query structure's biological unit, and the top hits will be structures that, at the most, match all four of the long proteins. Each of those hits will be listed as a "partial match."
- Pairwise alignments between individual protein components of the query structure and subject are compared to each other for compatibility, and compatible/matching alignments are clustered into sets of alignments that together constitute a biological unit match. Pairwise alignments are compatible: (1) if they do not share the same macromolecules, i.e., an individual protein molecule from the query structure structure cannot be aligned to two or more protein molecules from the subject at the same time; and (2) if they generate similar instructions (spatial transformation matrices) for the superpositions of co-ordinate sets. A simple distance metric can be used to compare transformation matrices, and it lends itself to cluster alignment sets efficiently.
- Filter by Taxonomy
 
- Filter results by domain (superkingdom) -- The "Filter by Taxonomy" options enable you to view the subset of similar structures from the "Eukaryota," "Bacteria," or "Archaea" domains (superkingdoms), from "Viruses," or from "Other" groups (such as synthetic constructs). Activate the check box(es) beside the group(s) of interest to see only the desired subset(s) of similar structures.
Note that a taxonomy filter will retrieve structures in which all biomolecules in the structure are from the selected superkingdom. For example, the "Viruses" filter will retrieve structures that only contain molecules from viruses, and not molecules from any other superkingdom. Structures that are composed of synthetic constructs, or composed of molecules from two or more superkingdoms, are included in the "Others" taxonomy category. For example, PDB ID 4N9F would be classified with a taxonomy of "Others" because it includes molecules from Human immunodeficiency Virus 1 and Homo sapiens, which fall into the superkingdoms of Viruses and Eukaryota, respectively.
|
 
The VAST+ search results page lists all structures that were found to be similar in 3D shape to the query.
Default sort order: The similar structures are shown as a ranked list, based on the following scores:
(1) number of aligned proteins, (2) RMSD, (3) number of aligned residues, and (4) sequence identity, in that order.
It is also possible to sort by taxonomy, if desired.
The blue arrow  that appears to the right of the column header indicates the primary sort criterion, and the direction (ascending/descending) of the sort. that appears to the right of the column header indicates the primary sort criterion, and the direction (ascending/descending) of the sort.
If desired, you can override the default sort order by clicking any arrow  beside a column header in the output table in order to choose that column as the primary sort criterion. In that case, your selected criterion is applied first, and the remaining sort criteria applied afterward (in their usual order). The arrow beside the selected column header will turn blue, indicating it is now the primary sort criterion, and indicating the direction of the sort (ascending or descending). beside a column header in the output table in order to choose that column as the primary sort criterion. In that case, your selected criterion is applied first, and the remaining sort criteria applied afterward (in their usual order). The arrow beside the selected column header will turn blue, indicating it is now the primary sort criterion, and indicating the direction of the sort (ascending or descending).
- Sort Options (Scores)
 
- Aligned proteins -- The number of protein molecules in the query structure have a match (i.e., similar 3D shape) to a protein molecule in the subject.
- RMSD -- The average root mean square deviation (RMSD), in Angstroms (Å), of all aligned residues between the query structure and the subject.
- This number is calculated after optimal superposition of two structures, as the square root of the mean square distances between equivalent C-alpha atoms. Smaller RMSD values indicate greater structural similarity. (Note that the RMSD value scales with the extent of the structural alignments and that this size must be taken into consideration when using RMSD as a descriptor of overall structural similarity.)
- The RMSD values displayed in VAST+ results might be much higher than those displayed in Original VAST results because the 3D alignment is optimized for the whole assembly rather than for individual protein molecules. Specifically, VAST+ takes all of the individual 3D domain alignments and calculates a global superposition. To do this, individual protein molecules might be shifted from their ideal pairwise superposition, in order to be able to generate a global superposition of all matched molecules between the query structure and the subject.
- For example, Original VAST might give RMSD values of about 3-4 Å at the worst, and rarely up to 6 Å for very large structures. In contrast, VAST+ results might generate more hits with RMSD values of 5-6 Å or more, and a number of cases with large values such as 20 Å or more. This can be due to flexibility in the biological unit, which might indicate something interesting. An example is the chaperone protein 3J1B: Cryo-em Structure of 8-fold Symmetric Ratcpn-alpha in APO State. The VAST+ neighbors for 3J1B include 3J03: Lidless Mm-cpn in the Closed State With Atpalfx, with an RMSD value of 13.41 Å.
- Aligned residues -- The total number of amino acids, from all protein molecules in the query structure's biological unit, that are spatially aligned to the subject.
- Sequence identity -- The percent of aligned residues that are identical, averaged over all of the aligned protein molecules (rounded to the nearest whole number on the VAST+ results display).
- Taxonomy -- Click on the up/down arrows in the taxonomy header to sort hits alphabetically by genus name.
If you choose to sort by taxonomy, the similar structures are sorted based on the following criteria:
(1) alphabetically by genus, then within a genus, the structures are sorted based on (2) number of aligned proteins, (3) RMSD, (4) number of aligned residues, and (5) sequence identity, in that order.
(Note: If you would like to only see the subset of search results from a specific superkingdom, use the taxonomy filter to select the desired superkingdom, then click on the arrows in the "Taxonomy" column header of the search results table to sort the structures from the superkingdom alphabetically by genus.)
- View details for a similar structure
 
-
Click on the plus button
 beside any structure to see a detailed view comparing the query structure to the matched structure. A separate section of this help document provides a description of the detailed view. beside any structure to see a detailed view comparing the query structure to the matched structure. A separate section of this help document provides a description of the detailed view.
- Search within results
 
- The "Search within results" text box enables you to enter a PDB ID, MMDB ID, or text word from the title of a structure of interest to find out if it among the similar structures retrieved by VAST, and if so, to instantly take you to the page of results on which it appears, with your structure of interest highlighted {in yellow}.
If you are viewing only a subset of the search results (for example, because you have checked a box beside a bar graph of interest to filter the results), and your structure of interest is not in that subset but is in the overall search results, VAST will display a relevant message that includes a link which opens the complete set of search results, automatically highlighting your structure of interest.
If, on the other hand, you enter a PDB ID of a structure that is not listed among the VAST results, you will see an error message indicating that your structure of interest was not among the results.
- Example: Retrieve the VAST results for 1HBB: "High-resolution X-ray Study of Deoxyhemoglobin Rothschild 37beta Trp-> Arg: a Mutation That Creates an Intersubunit Chloride-binding Site (human hemoglobin)." Let's say that you suspect 1UMO ("The Crystal Structure Of Cytoglobin: The Fourth Globin Type Discovered In Man") will be one of the structures retrieve and you quickly want to find where that structure falls within the ranked list of similar structures. To do that, simply enter the PDB ID 1UMO (or its corresponding MMDB ID 66087) in the "search within results" text box, and you will be taken immediately to the appropriate page of search results, with 1UMO highlighted in yellow. You can then click on the plus button
 to the left of that structure in order to see more details about the comparison between 1HBB and 1UMO. If you enter the PDB ID or MMDB ID of a structure that is not similar (as determined by VAST+) to the query structure, a pop-up message will appear that says, "XXX is not a simlar structure of 1HBB." to the left of that structure in order to see more details about the comparison between 1HBB and 1UMO. If you enter the PDB ID or MMDB ID of a structure that is not similar (as determined by VAST+) to the query structure, a pop-up message will appear that says, "XXX is not a simlar structure of 1HBB."
|
(on the search results page)  
An "Original VAST" button is located near the top of a VAST+ search results pageIt gives you the option to view your search results in the original style VAST display, which lists structures that have similarities to individual protein molecules or individual 3D domains in your query structure, and allows you to view their sequence alignments and 3D superpositions. An illustrated example of the original-style VAST results is provided in the Graphical Displays section of this document.
(In contrast, the new VAST+ search results rank similar structures based on their biological unit similarity to the query structure (illustrated examples of VAST+ results), and provide the ability to view sequence alignments and superpositions of the biological units)
A separate section of this document describes the difference between Original VAST and VAST+.
|
 
|
VAST+ results are available for most, but not all, structures. If your query generates the message, "No matched structure found," it can be for one of the reasons listed in the table below. If your query does not have VAST+ neighbors, but does have Original VAST neighbors, follow the link provided in the error message to view the available results. If your query structure is relatively new, try your search again at a later date, as VAST (and VAST+) neighbors are calculated/updated weekly.
|
| Circumstances under which a structure has neither VAST+ nor Original VAST results: |
| VAST neighbors not yet calculated |
VAST neighbors are currently being calculated for your query structure and are therefore not yet available. Try your search again in several days, as VAST (and VAST+) neighbors are calculated/updated on a weekly basis. |
| structure is too small |
In order to be processed by VAST, a protein molecule must have at least three secondary structure elements (alpha helices and/or beta sheets). If an individual protein in a structure contains only one or two secondary structure elements, that protein will not be processed by VAST. Structures that are composed entirely of such proteins will therefore have no VAST neighbors. |
| structure has no protein |
Currently, VAST works only on protein molecules. If a structure does not contain any proteins, for example, if it contains only DNA or RNA molecules, it will not be processed by VAST, and will therefore have no VAST neighbors. |
| structure is unique |
The structure is large enough to be processed by VAST+ (i.e., has at least three secondary structure elements), but has a unique 3D shape which has not been found among any of the other publicly available structures. |
| Circumstances under which a structure has no VAST+ results, but does have Original VAST results: |
| VAST+ neighbors not yet calculated |
VAST+ neighbors are currently being calculated for your query structure and are therefore not yet available. As noted in the overview section of this document, the VAST+ is an extension of, and built upon, the original VAST system. Therefore, there may be some lag between the availability of original VAST data and the subsequent appearance of VAST+ data in the system. Try your search again in several days, as VAST (and VAST+) neighbors are calculated/updated on a weekly basis.
|
| structure has no biological unit defined |
If a structure record does not have a biological unit defined in its source PDB file, it is only available as an asymmetric unit (i.e., raw structure data). In that case, the structure is not processed by VAST+, since VAST+ focuses on finding similarities among biological units. However, such structures are processed by the Original VAST algorithm, which may have found structures that are geometrically similar to individual proteins, or individual 3D domains, in your query structure. |
| merged structure |
Merged structures, which have been assembled from PDB split files, do not have a biological unit defined, and are therefore not processed by VAST+. However, such structures are processed by the original VAST algorithm, which may have found structures that are geometrically similar to individual proteins in your query structure. In that case, view the original VAST results for your query structure. |
| structure is too large |
Structures that have biological units composed of 60 or more protein molecules are not currently processed by VAST+. However, such structures are processed by the original VAST algorithm, which may have found structures that are geometrically similar to individual proteins in your query structure. In that case, view the original VAST results for your query structure. |
structure has no full length
protein neighbor |
The structure has been neighbored by VAST, but did not generate VAST results for the any full-length protein molecule in the query, so the structure has not been considered for VAST+.
VAST+ results only list structures that have a complete match or partial match to your query structure's biological unit, or have a match to at least one full protein molecule in the query structure.
Even if no full length protein matches are found, the query structure might still have hits in Original VAST, because it is still possible that a portion of a protein in the query structure, or an individual 3D domain, has a match to another structure. In that case, view the original VAST results for your query structure. |
(on the search results page)  
To retrieve the VAST+ results for a different structure, simply enter its PDB ID or MMDB ID in the text box in the upper right hand corner of a VAST+ results page and press the "New Search" button. Or, enter either ID on the VAST+ query page.
|
|