NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.
This publication is provided for historical reference only and the information may be out of date.
Scope
NCBI’s Gene database is designed to aggregate gene-specific information from multiple perspectives including sequence, mapping, publications, function of products, expression, evolution, and consequences of variation. Gene makes these data available for diverse scenarios, from occasional interactive access on the Web through computational access to selected or complete data sets.
Gene assigns an identifier (the GeneID) for each gene in each taxon either represented in the NCBI Reference Sequence (RefSeq) project, or under consideration by RefSeq. Usually this taxon is defined at the species level, but sometimes will be per isolate, strain or cultivar. Gene is closely coupled with RefSeq, in that genes annotated on RefSeq sequences are assigned GeneIDs for tracking. Not all records in Gene, however, are based on RefSeqs. Gene works closely with multiple groups that may identify a gene before it has been been defined by sequence. In other words, some records in Gene are mapped traits or other phenotypes.
This document does not provide detailed instructions about how to use Gene or comprehensive details about how Gene is built from numerous data sources. For detailed, up- to-date documentation, please refer to Gene’s Help document.
History
The database currently known as Gene was first made public in 1999 as LocusLink (1). There was only one species represented (human) and little more than 9000 records. The Web interface supported links only to dbSNP, OMIM, RefSeq, GenBank, and UniGene within NCBI, as well as to the now defunct Genome Database (GDB) and a few other databases externally (Figure 1). By late 2003, when Entrez Gene was released, there were 10 species, almost 195000 records, and links computed to dbSNP, Ensembl, the HUGO Gene Nomenclature Committee (HGNC), GEO, Map Viewer, Mammalian Gene Collection (MGC), Nucleotide, Protein, PubMed, Taxonomy, UCSC, UniSTS, UniGene, and multiple species-specific model organism databases (Figure 2). Now Gene represents more than 11,000 taxa, more than 13,000,000 records, and more than 40 types of links to other NCBI databases.
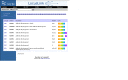
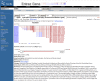
Figure 1.
Representation of gene-specific information in LocusLink.
In addition to the taxonomic scope, the number of records, and connectivity, the look and feel of Gene has changed over the years. The current database implementation provides a hierarchical Table of Contents to facilitate navigation, integration with MyNCBI to support personalized display of sections of the record, an embedded viewer of NCBI’s annotation of any gene on one or more genomic RefSeqs, a page dedicated to the display of GeneRIFs, and, especially for human, enhanced access to gene-specific variation and phenotype reports (Figure 3).
Data Model
Gene has a simple data model. Once the concept of a gene is defined by sequence or mapped location, it is assigned a unique integer identifier or GeneID. Then data of particular types are connected to that identifier. These types include sequence accessions, names, summary descriptions, genomic locations, terms from the Gene Ontology Consortium (2), interactions, related phenotypes, and summaries of orthology. For some of the commonly requested elements, and because of the simplicity of the data model, Gene provides tab-delimited files of content anchored on the GeneID.
The full extraction of Gene is exported as binary ASN.1 (ftp://ftp.ncbi.nih.gov/gene/DATA/ASN_BINARY/) with a tool provided to convert to XML (ftp://ftp.ncbi.nlm.nih.gov/asn1-converters/by_program/gene2xml). The ASN.1 representation of a Gene record (http://www.ncbi.nlm.nih.gov/IEB/ToolBox/CPP_DOC/lxr/source/src/objects/entrezgene/entrezgene.asn) incorporates several objects used by other resources (Gene-ref, BioSource, RNA-ref, etc.), but also has several several objects specific to Gene to represent the type of gene, map location, and properties. A major component of Gene’s ASN.1 representation is the generic Gene-commentary that is used to represent content defined by type, heading, label, text, and source.
Dataflow
Gene is updated daily and incrementally. In other words, on any given day a record may be changed but not all records will be changed on the same day. The FTP site is refreshed comprehensively each day, except for special reports and documentation files.
Data are added to Gene by integrating automated and curatorial flows. For some taxa, primarily genomes submitted to NCBI with annotation of genes, data are loaded to Gene by extracting information annotated on the gene feature of the genomic sequence that was submitted. Those data may be supplemented by data from Gene Ontology (GO) based on identifiers in the sequence, according to rules reported by Gene in ftp://ftp.ncbi.nih.gov/gene/DATA/go_process.xml. The content of the Gene record for these species is thus updated only when a new annotation of the genome is supplied, or when supplementary data such as GeneRIFs, GO terms, or UniGene clusters are updated.
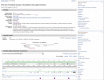
For the taxa included in RefSeq’s curated set (see the RefSeq chapter for more information), updates may happen daily, and independently of a re-annotation of a genome. There are automated flows to reconcile official gene symbols and full names, protein names, and database identifiers. Curators may modify summaries, add or redefine transcript or RefSeqGene RefSeqs, or add citations to the record. When this happens the Gene record is updated. Genes that are in scope for more frequent updates can usually be detected because the NCBI Reference Sequences section will include a subsection entitled RefSeqs maintained independently of Annotated Genomes (Figure 4).

Figure 4.
A record maintained independently of annotation releases. If this information is included in the Reference Sequences portion of the Gene record, other content of the record is also likely to change more often.
More detailed information about the maintenance of information in Gene is provided in Gene Help.
Access
Web
Gene
Gene is accessed on the Web via http://www.ncbi.nlm.nih.gov/gene/. If the GeneID is known, the path to a specfic record is generated based on the root path plus the GeneID, e.g., http://www.ncbi.nlm.nih.gov/gene/672 for human BRCA1, for which the GeneID is 672.
Gene’s website is searched via NCBI’s Entrez system. The fields, filters, and properties that support effective queries are documented in Gene’s Help book. Among those that are used most often are the gene symbol ([gene]) and a sequence accession.
Other NCBI databases
Gene is also accessed from other databases at NCBI. For example, a query to sequence databases, ClinVar, MedGen, or PubMed will detect what looks like a Gene symbol, and provide a display summarizing what is available in Gene (Figure 5). Records in Gene related to other database entries can be identified by following the links to Gene displayed in the panel at the right.

Figure 5.
Gene sensor in PubMed. A query that matches a gene symbol provides the user with link to more information in Gene, as well as the listing of citations in PubMed that satisfy the query (http://www.ncbi.nlm.nih.gov/pubmed/?term=pcdhga12)
FTP
Information about genes is accessible from any FTP site of NCBI that includes GeneIDs as part of the content. These will not be enumerated in this document; just be aware that if a record reports a GeneID or gene_id, that is the identifier from NCBI’s Gene database.
Gene
Gene’s FTP site (ftp://ftp.ncbi.nlm.nih.gov/gene/) is divided into DATA, GeneRIF, and tools sections. The ftp://ftp.ncbi.nlm.nih.gov/gene/README file describes all sections, reports maintenance details, and provides detailed information about files available from Gene, as well as the annotation-specific files provided from ftp://ftp.ncbi.nih.gov/genomes. The DATA subdirectory provides several comprehensive files, but also includes subdirectories for the full extractions (ASN_BINARY) and tab-delimited reports (GENE_INFO) that provide subsets of data divided by major taxonomic groups.
GFF
For those interested in the location of genes and exons in a genomic context, the genomes path provides a GFF directory for many species. The README_CURRENT_RELEASE file indicates the NCBI Annotation Release being reported and the dates on which data were frozen to support the annotation. In the GFF file, GeneID is reported as a cross-reference, e.g., Dbxref=GeneID:1080. NCBI uses the GFF3 standard.
E-Utilities
Gene is fully accessible programmatically using NCBI’s E-Utilities. The tools section on Gene’s FTP site provides some sample perl scripts to extract information from Gene based on esummary and efetch and elink.
Related Tools
In addition to the scripts available from the tools directory of Gene’s FTP site (ftp://ftp.ncbi.nlm.nih.gov/gene/tools/README), gene2xml (ftp://ftp.ncbi.nlm.nih.gov/asn1-converters/by_program/gene2xml/) supports conversion of Gene’s ASN.1 representation to XML. Gene-related programming tips are included in Gene Help and FAQ.
References
- 1.
- Maglott DR, Katz KS, Sicotte H, Pruitt KD. NCBI's LocusLink and RefSeq.Nucleic Acids Res.2000Jan 12811268 [PMC free article: PMC102393] [PubMed: 10592200]
- 2.
- Gene Ontology ConsortiumBlake JA, Dolan M, Drabkin H, et alGene Ontology annotations and resources.Nucleic Acids Res.2013Jan41Database issueD5305doiEpub 2012 Nov 1710.1093/nar/gks1050 [PMC free article: PMC3531070] [PubMed: 23161678] [CrossRef]
- Gene - The NCBI HandbookGene - The NCBI Handbook
- Chromosome neighbors for GEO Profiles (Select 108576356) (20)GEO Profiles
- Chromosome neighbors for GEO Profiles (Select 108582587) (18)GEO Profiles
- Concise Conserved Domain Links for Protein (Select 1829777429) (1)Conserved Domains
- ARHGAP33 Rho GTPase activating protein 33 [Homo sapiens]ARHGAP33 Rho GTPase activating protein 33 [Homo sapiens]Gene ID:115703Gene
Your browsing activity is empty.
Activity recording is turned off.
See more...