Making Submission in SRA Submission Portal
Introduction
![]() The Submission Portal Wizard provides a browser-based user interface for submitting SRA metadata as well as various options for uploading data files.
The Submission Portal Wizard provides a browser-based user interface for submitting SRA metadata as well as various options for uploading data files.
![]() For better understanding of SRA submission process and SRA data organization please refer to SRA Metadata and Submission Overview and see our 'How to Submit' video tutorial.
For better understanding of SRA submission process and SRA data organization please refer to SRA Metadata and Submission Overview and see our 'How to Submit' video tutorial.
Prerequisites
- SRA accepts reads from high throughput sequencing platforms in specific formats (see the SRA File Format Guide for details). SRA does NOT accept assembled/consensus data or contigs.
- For submissions of 1000 samples or less. If you have more than 1000 samples, please create multiple submissions with the same BioProject reference.
- Files can be compressed using gzip or bzip2, and may be packaged in a tarball. Do not use zip! Packaging and/or compressing your files is not required.
- Each submitted fastq file should be less than 100GB in size. If compressed files are larger than 100GB, please split them prior to submission.
- Studies with more than 5TB of data should be split across multiple submissions, keeping each set of uploads under 5TB, and waiting for completion of each submission before uploading the next set of files. Submissions can be linked to the same BioProject to ensure all data are searchable with a single accession.
- SRA does not accept submission of duplicate files and these submissions may be suppressed without warning. In addition, submitting duplicate data will lead to significant delays in processing of the submission. To update an existing record, do not resubmit the data files, instead contact the SRA helpdesk (sra@ncbi.nlm.nih.gov) for assistance.
Submitting
Log in to the SRA Submission Portal Wizard
On the Submission Portal Home Page https://submit.ncbi.nlm.nih.gov/ navigate to SRA or log in to the SRA Submission Portal Wizard directly.
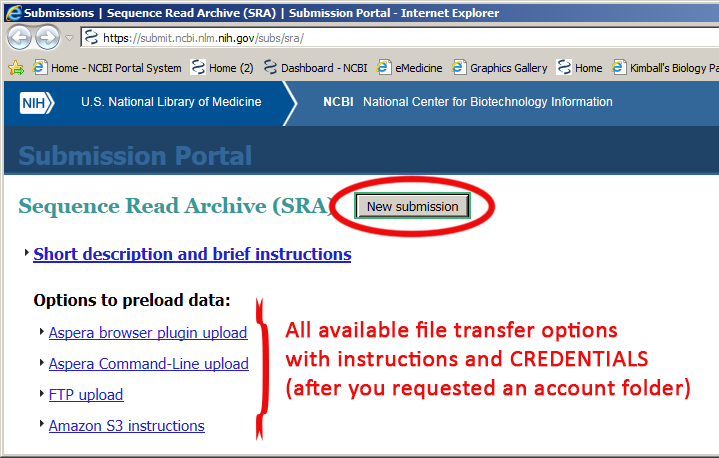
Create new submission
Request a personal account folder to pre-upload your sequence data files (for the first time users) by clicking on the button .
Figure 1: Request personal account folder for file pre-upload

Requesting the account folder is recommended to allow for greater flexibility in file uploads. If you skip this step here, you will be given an opportunity to request the personal account folder later on the Files step of the submission.
Click the button :
Figure 2: Create new SRA submission
Follow steps
Figure 3: SRA submission's steps including registration of new BioProject and BioSamples

You may leave your submission at any step and return to finish it later.
1. Submitter
- Check accuracy of your contact information and update it if necessary.
- Create a group of collaborators if necessary.
- Plan to use your primary email address in your communications with SRA staff.
- Press the button .
2. General Info
Select Yes in the first two sections of this step if you already registered your project and samples at the BioProject and BioSample Submission Portal Wizards. If you select No, the SRA Submission Wizard will ask you to create them, which may result in creating duplicates of your existing project and/or samples and this will lead to submission errors.
Provide the release date in the third section of this step. The default release date is set to Release immediately following processing.
You will be able to modify your release date later in the SRA User Interface.
3. Project Info
Please refer to the SRA BioProject & Biosample Guide.
![]() In the SRA Submission Portal Wizard, registration
of BioProject is simplified. The BioProject accession (
In the SRA Submission Portal Wizard, registration
of BioProject is simplified. The BioProject accession (PRJNA#) is assigned after
the SRA submission is finished and is not required for SRA metadata.
Please read SRA specific guidelines here: BioProject & BioSample.
4. BioSample type
Select a package for your samples. Please refer to BioSample Packages.
If you determined that your human data must be submitted via dbGaP, please delete your current submission and refer to dbGaP Submission Guide.
5. BioSample attributes
Please do not use BioSamples that were registered in another user's account.
Select the most appropriate taxon for each organism/metagenome in the NCBI Taxonomy Browser. Providing the exact and most specific name for the organism/metagenome will simplify your BioSample submission.
For more information refer to BioSample documentation.
Refer to SRA Troubleshooting Guide to resolve common errors or warnings that occurred on this step.
6. SRA Metadata
There are two options for providing the SRA metadata:
- Use built-in editor (sutable for submissions with fewer samples and provides some of required validation on the fly)
- Upload a file (more convenient for large submissions)
Figure 4 - Built-in editor

If you decided to upload a file with your metadata, you need to download the Excel spreadsheet template from the SRA metadata page.
Instructions for metadata
-
If you just created project in the SRA wizard, leave the column bioproject_accession empty (if the column is present).
-
If you created samples previously, provide accessions in the form of SAMN# in the column sample_accession.
-
If you created samples in the SRA wizard, provide names of samples that you just created in the column sample_name. Please note that sample names must be unique within your entire account.
-
Each row in the template represents an EXPERIMENT. EXPERIMENT is a unique combination of sample + library + sequencing strategy + layout + instrument model. Each experiment should have a unique library_ID that is short and meaningful (like an ID you might use in lab).
-
Only one RUN per EXPERIMENT is allowed in the Submission Portal. When libraries are indeed identical (same combination of sample + library + strategy + layout + instrument model), all files should be placed in the same RUN. To do this simply enter the file names consecutively in the same row. PAIRED files must always be listed in the same row.
-
If uploading a bam file of reads aligned against a published NCBI assembly then enter the NCBI assembly name in the Reference assembly column. If your bam reads were aligned against a non-NCBI assembly then enter the filename of the custom reference in the Reference assembly column. Upload your non-NCBI assembly in fasta format (please do not compress the fasta file).
-
When submitting unaligned bam files (eg: PacBio
*subreads.bam) please enter unaligned in the Reference assembly column. -
If you archived your files with the tar utility, you must list the individual file names that are contained in the archive on your SRA metadata sheet (the one exception is a tar of .fast5 files for Nanopore native file submissions).
-
Provide exact file names (including extensions) in the filename columns. If uploading more than 8 files per sequencing library/run, please use the Excel spreadsheet instead of the embedded sheet as more filename# columns can be added to the Excel spreadsheet template as needed.
-
File names must be unique within a submission.
Click the button .
Refer to SRA Troubleshooting Guide to resolve common errors or warnings that occurred on this step.
7. Files
Select one of the upload methods:
- I have all files preloaded for this submission (best method for all submissions)
- I will upload all the files now via HTTP/Aspera (suitable for submissions with a few small files)
Figure 5

After you requested your personal account folder (either before starting your submission or on this step) AND checked the radiobutton "I have all files preloaded for this submission", the instructions and credentials will be provided on the page.
Detailed FTP and Aspera instructions can also be found here: SRA File Upload.
After you uploaded your files to your submission folder as instructed, please wait 10-15 minutes and click on the button .
Figure 6

After you finished uploading your files via Aspera/HTTP or selecting your submission folder, click the button .
Refer to SRA Troubleshooting Guide to resolve common errors or warnings that occurred on this step.
8. Overview
Please review your submission's summary and make sure that everything is correct including the release date.
At this stage, you can return to any step of your submission by clicking on the corresponding tabs.
.
![]() Once submitted, your submission is queued
for processing. Allow at least 24 hours after submitting to receive feedback from SRA before contacting us. Note that if you created
a BioProject or/and a BioSample submissions within the SRA Wizard, you will receive feedback from these databases first.
Once submitted, your submission is queued
for processing. Allow at least 24 hours after submitting to receive feedback from SRA before contacting us. Note that if you created
a BioProject or/and a BioSample submissions within the SRA Wizard, you will receive feedback from these databases first.
Downloading accessions
All your SRA submissions can be viewed on the SRA Submission Portal's home page: https://submit.ncbi.nlm.nih.gov/subs/sra/.
Figure 7

Your submission's Overview page can be found by clicking on the link SUB# in the column Submission or by updating and entering the below link in your address bar:
https://submit.ncbi.nlm.nih.gov/subs/sra/[insert your SUB# here]/overview
Troubleshooting
For help with the SRA Submission Wizard refer to SRA Submission Portal Troubleshooting Guide.
Updating
Refer to the Update Guide: Updating SRA data.
Contact SRA staff
Before contacting SRA staff for assistance please visit our SRA Submission Portal Troubleshooting Guide.
Email sra@ncbi.nlm.nih.gov for help.
![]() Please provide your submission's temporary ID in the form of
Please provide your submission's temporary ID in the form of SUB# in your messages.
