Web APIs
How to use
Publication
Chih-Hsuan Wei, Robert Leaman, Zhiyong Lu (2016). Beyond accuracy: Creating interoperable and s calable text mining web services, Bioinformatics, DOI:10.1093/bioinformatics/btv760.
Introduction
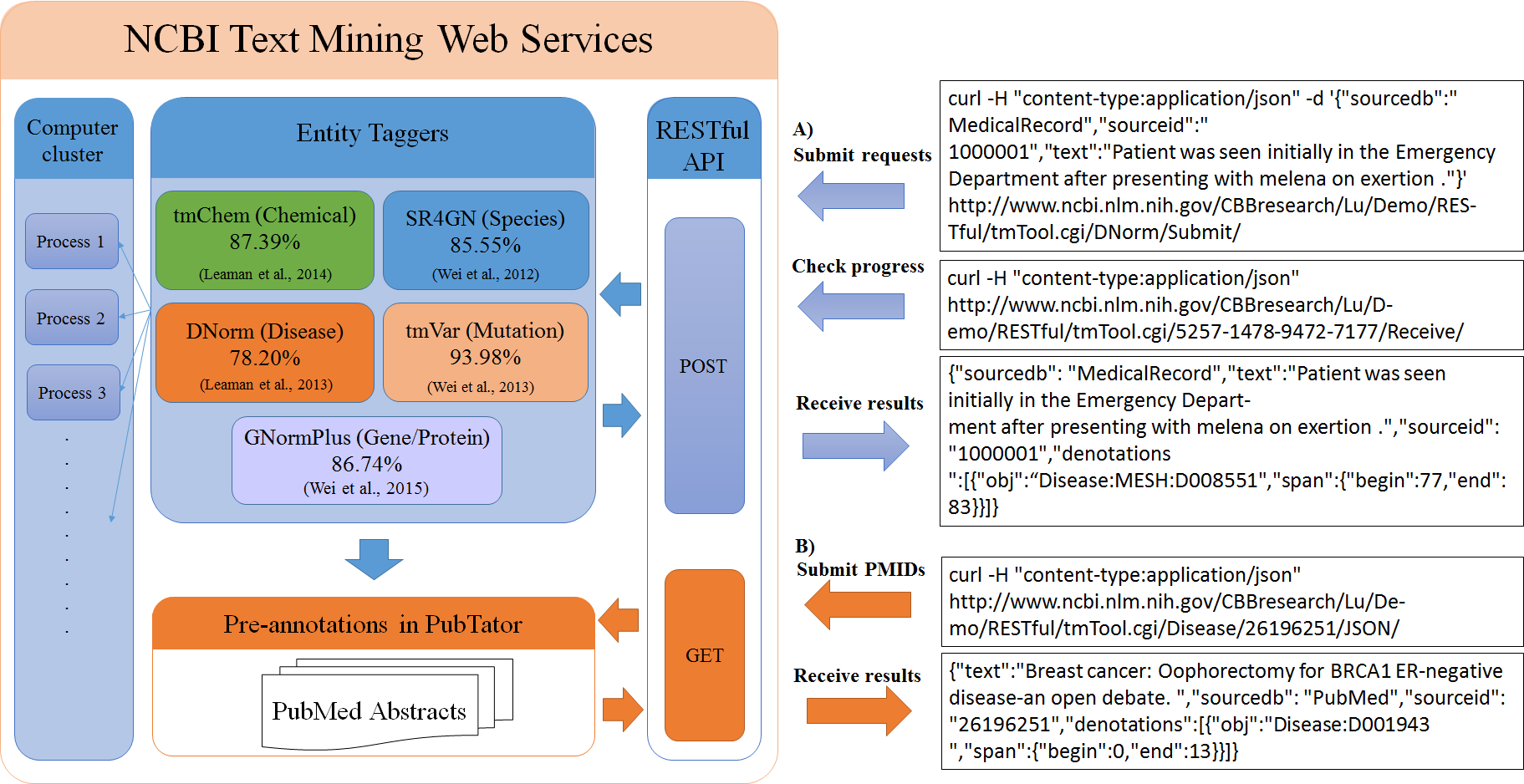
We report here our recently developed web-based text mining services for biomedical concept recognition and normalization. The below Figure describes the overall architecture of our web services, which use sta ndard HTTP method calls (often known as RESTful services) and allow two access modes: A) a batch-oriented processing function for any arbitrary text input (abstract, full text, patent, etc), submitted via HTTP POST an d B) instant retrieval of pre-tagged results of PubMed abstracts via HTTP GET. For the batch processing function, users may submit one or multiple documents per batch, and large requests will be sent to a computer clu ster for parallel processing. When retrieving pre-tagged results of PubMed abstracts, the request only requires the PMIDs of the requested abstracts. This option is provided because annotating biomedical literature is the most common use case for such a text-mining service. From a technical stand-point, the preprocessing is made possible by our previous system PubTator, which stores text-mined annotations for every article in PubM ed and keeps in sync with PubMed via nightly updates.
Performance
| Taggers | Bioconcepts | Evaluation corpus | Precision | Recall | F-measure |
| GNormPlus (Wei et al., 2015) | Gene | BioCreative II - GN | 87.08% | 86.41% | 86.74% |
| tmChem (Leaman et al., 2014) | Chemical | CHEMDNER | 89.09% | 85.75% | 87.39% |
| DNorm (Leaman et al., 2013) | Disease | NCBI Disease corpus | 80.30% | 76.30% | 78.20% |
| tmVar (Wei et al., 2013) | Mutation | MutationFinder | 98.80% | 89.62% | 93.98% |
| SR4GN (Wei et al., 2012) | Species | Linnaeus corpus | 85.82% | 85.28% | 85.55% |
| Taggers | HTTP Submission Method | Description | Throughput (seconds per article; averaged over 5000 articles) |
| PubTator | GET | Return Pre-Annotations |
0.044s |
| GNormPlus* | POST | On-Demand Processing | 0.127s |
| tmChem | POST | 0.008s | |
| DNorm | POST | 0.007s | |
| tmVar | POST | 0.090s |
* SR4GN is used by GNormPlus. The results of GNormPlus include both gene and species.
Supplementary
We provide below several sample codes to show how to use our RESTful API service via
programs.
RESTful sample codes avaliable in Perl, Python and Java.
We also provide a script for input formats (i.e., BioC, PubTator and JSON) checking.