NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter pylori: Physiology and Genetics. Washington (DC): ASM Press; 2001.
Cell-envelope components are the first points of contact between bacterial pathogens and the host. Many of the distinguishing properties of Helicobacter pylori are related to the constituents of this cellular compartment. This chapter reviews the physical and biochemical composition of the H. pylori cell envelope, with reference where appropriate to the microbiological and pathogenic implications.
The H. pylori cell has been shown to have an overall relatively hydrophilic and negatively charged surface in vitro (91). The outer membrane composition is unique in its protein content and lipopolysaccharide structure (4, 5, 71, 72). However, the physical organization of the cell envelope is similar to that of other gram-negative bacteria. Difficulty has been reported in separating inner and outer membranes (21), similar to the case with Campylobacter sp. (82). This is due to extensive linkages between the membranes, possibly facilitated by the numerous lipoproteins identified in the H. pylori genome (99). Procedures have, however, been established for separating individual membrane fractions (21) and outer membrane proteins (42, 108).
Peptidoglycan
The peptidoglycan of H. pylori differs substantially from that of Escherichia coli (14). H. pylori peptidoglycan contains a high proportion of muropeptides, with a pentapeptide side chain ending in glycine and containing (1–6)-anhydro-N-acetylmuramic acid. It lacks murein-bound lipoprotein, trimeric muropeptides, and (l-d) cross-linked muropeptides. In coccoid cells, activation of a (γ)-glutamyl-diaminopimelate endopeptidase leads to massive conversion of tri- and tetrapeptide monomers into dipeptide monomers (14). This has also been observed for sporulating Bacillus sphaericus (102). The H. pylori genome has homologs of all the enzymes required for the cytoplasmic synthesis of precursors required for peptidoglycan assembly (19, 99). After transport through the cytoplasmic membrane, peptidoglycan precursors are incorporated into the peptidoglycan layer by the penicillin-binding proteins (PBPs). It was originally suggested that H. pylori produces four PBPs (51, 64). It was inferred from consideration of the genome sequence that three have homology to known PBPs (99) and a fourth (PBP 4; TIGRHP0160) has no sequence similarity to any known PBP or proteins from other bacteria (64). However, a recent study using labeled ampicillin identified eight PBPs (44). When these authors scrutinized the putative PBPs encoded by the genome sequence for characteristic motifs, none possessed all three motifs found in conventional PBPs, suggesting that the PBPs of H. pylori are unique (44).
Fatty Acid and Lipid Composition of H. pylori
The unusual cellular fatty acid profile of H. pylori was a major criterion for the exclusion of the organism from the genus Campylobacter (40). The fatty acid compositions of four strains of H. pylori have been analyzed by gas-liquid chromatography (39). The fatty acids identified were myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), 19-carbon cyclopropane fatty acid (C19:0cyc), β-hydroxy-palmitic acid (3-OH-C16:0), and β-hydroxystearic acid (3-OH-C18:0). The most abundant cellular fatty acids were myristic acid (31 to 45%) and 19-carbon cyclopropane fatty acid (20 to 24%). All other fatty acids were found in amounts of 12% or less of total cellular fatty acids. Myristic acid (41 to 55%) and 19-carbon cyclopropane fatty acid (22 to 30%) were the major phospholipid fatty acids detected, while β-hydroxy fatty acids and unsaturated fatty acids were either not detected or present in very small amounts.
The H. pylori total lipid content (by weight) is 6% neutral lipids, 20.6% glycolipids, and 73.4% phospholipids (46). The major phospholids are phosphatidylethanolamine, cardiolipin, and phosphatidylglycerol. Phosphatidylserine was detected as a minor phospholipid. Three kinds of cholesterol glucosides accounting for about 25% (wt/wt) of total lipid have also been described (46). Cholesterol glucosides are very rare in animals and bacteria, but they have been found in 13 out of 15 Helicobacter species examined (43). Thus, the presence of cholesterol glucosides is a unique characteristic feature of the Helicobacter sp. membrane.
Lipopolysaccharide
A key component of the outer membrane of H. pylori is lipopolysaccharide (LPS). Fresh clinical isolates of H. pylori produce high-molecular-weight smooth-form LPS (S-LPS), which consists of an O side chain, a core oligosaccharide, and lipid A. However, strains of H. pylori that have been repeatedly subcultured on solid media produce low-molecular-weight rough-form LPS (R-LPS) that lacks the O side chain. Strains producing S-LPS can revert and produce R-LPS when they are grown in liquid media (105). A compositional analysis of H. pylori R-LPS isolated from three different culture collection strains of H. pylori found that the core oligosaccharide contains fucose, d-mannose, d-glucose, d-galactose, d-glycero-d-manno-heptose, l-glycero-d-manno-heptose, and 3-deoxy-d-manno-2-octulosonic acid (73). The molar ratio of hexoses differed between different strains, reflecting structural differences. A more recent study, however, failed to find mannose as a constituent (4). LPS is treated extensively elsewhere in this volume (see chapter 8).
Lipid A
The lipid A component of H. pylori LPS is unusual in that it has low mitogenic and pyrogenic activities and low lethal toxicity compared with lipid A from other enterobacteria (74). The chemical structure of the lipid A component of H. pylori R-LPS and S-LPS has been elucidated (73). The hydrophilic backbone of the molecules consisted of β-(1,6)-linked d-glucosamine (GlcN) disaccharide 1-phosphate. In S-LPS, the hydroxyl group at position 4 was partially substituted by a phosphate group. It is likely that the distinctive phosphorylation and acylation patterns in H. pylori lipid A combine to produce low biological activity.
O Antigen
The most striking feature of the O antigen is the presence of extended chains with fucosylated and nonfucosylated N-acetyllactosamine units. The repeating units of the O side chains of LPS of H. pylori strains have been shown to mimic type 2 Lewis blood group antigens (Lex and Ley) in structure (5). Extensive serological and structural studies have recently shown that H. pylori strains may also carry type 1 blood group determinants, namely, Lea, H-1 (Led), and the type 1 chain precursor Lec (71). Many α-2-fucosyltransferases are involved in the addition of fucose to create the A, B, H, and Lewis blood group antigens. These include the products of the H, Se, A, B, X, and Le genes. In the case of Lea and Leb antigens, Lea is created by fucosylation of the type 1 precursor by the product of the Le gene whereas Leb is created by the fucosylation of the type 1 precursor to form the H type 1 antigen followed by fucosylation by the product of the Le gene to give the Leb antigen. A similar pathway is used in the biosynthesis of the Lex and Ley blood group antigens, except both are formed from the type 2 precursor. The type 2 precursor is fucosylated by either the product of the Le or X genes forming Lex or the precursor is fucosylated by either the Se or H gene product to form the H type 2 antigen and then fucosylated further by either the Le or X gene product to form Ley. Thus, the pathway is made up of various enzymes competing for the same substrate (Fig. 1). H. pylori may use similar enzymes to synthesize Lewis antigens. The genome of H. pylori strain 26695 contains two copies of an α1,3-fucosyltransferase (37, 99). Recently, an α1,2-fucosyltransferase (106) and a β1,4-galactosyltransferase (66) have also been described.
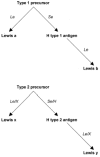
Figure 1
The biosynthesis of the Lewis blood group antigens. In italics: the transferases involved in each step of biosynthesis. In bold: the blood group antigens formed after each step.
Phase Variation
Many strains express Lex and Ley (89), but only some have been shown to express type 1 epitopes (71, 107). H. pylori LPS has been shown to display phase variation (3). From a single strain, various serotypes were isolated, all expressing different LPS structures and having different glycosyltransferase levels, which demonstrates that in H. pylori, phase variation contributes to increased heterogeneity. A subsequent study showed that length variation of poly(C) tracts in the α3-fucosyltransferase genes causes reversible frameshifting and disruption of the open reading frame. The on-off status of the genes governs the LPS serotypes of phase variants and clinical isolates (2).
Qualitative differences in LPS profiles are observed between H. pylori cells grown at pH 5 or pH 7, which suggests that H. pylori may alter its LPS structure in response to acidic pH (69). This may be an important adaptation facilitating H. pylori colonization of the acidic gastric environment (chapter 8).
Role of LPS in Colonization and Pathogenesis
Purified LPS inhibits the binding of H. pylori to laminin, indicating that LPS is involved in the adhesion process (103). The Lex antigen has been shown to mediate adhesion of H. pylori to fixed sections of gastric tissue (28). Mutational analysis of the gene encoding the β-1,4-galactosyltransferase demonstrated that this enzyme plays a key role in the biosynthesis of the polysaccharide O chain backbone by catalyzing the addition of Gal to GlcNAc. A mutated strain that produced truncated LPS colonized mice less efficiently than the parental strain, indicating that LPS plays an important role in colonization (66).
H. pylori LPS stimulates basal and gastrin-stimulated histamine secretion in rat enterochromaffin-like cells (59) and stimulates proliferation of the same cells. This property of LPS may be important in vivo as infected individuals have been reported to have a higher acid output than noninfected controls (49).
Surface Localization of Cytoplasmic Proteins
The cell surface of H. pylori has the unusual property of being able to incorporate proteins such as urease, catalase, HspA, HspB, and superoxide dismutase (SOD), which are found virtually exclusively within the cytoplasm in other bacteria (8, 23, 45, 87, 93). Urease, catalase, and SOD are discussed in detail elsewhere in this volume (chapters 15 and 16). Cryo-immunolocalization techniques have demonstrated that urease, catalase, and HspB are located strictly within the cytoplasm of freshly subcultured, early log-phase H. pylori. However, at the end of the log-phase, these proteins are also surface associated or extracellular (85). Significant fractions of urease and HspB are also surface associated in vivo (24). Indirect gold immunostaining of H. pylori SOD with a polyclonal antibody directed against the iron-containing SOD of E. coli showed a surface localization of the enzyme (93). Gold particles were distributed on the cell envelope and on the sheath of the flagella. SOD was located on the outer surface of a limited number of bacteria, but the enzyme was cytoplasmic in most cells.
Mechanism of Surface Localization of Cytoplasmic Proteins
In contrast to other H. pylori proteins with specific secretion pathways (10, 97), the mechanism whereby urease, HspB, catalase, and SOD become associated with the other membrane of H. pylori is controversial. Urease and HspB are large oligomeric proteins (6) that are typically found exclusively in the cytoplasm of bacteria and would not be expected to cross the bacterial outer membrane. H. pylori urease, SOD, and catalase have been shown genetically to lack leader peptides (18, 34, 78, 93, 96). Alternative transport mechanisms must therefore exist. Some very elegant work has shown that although H. pylori urease, HspB, and catalase are intrinsic cytoplasmic proteins in log-phase bacteria, these proteins become associated with the outer membrane of H. pylori when they are released from the cytoplasm by autolysis of a fraction of the bacteria (85). They then become adsorbed to the surface of intact bacteria. Scott et al. (88) also reported that the major source of external urease originated from lysis of the organism. However, Vanet and Labigne (104) used a subcellular fractionation and quantitative Western blot approach to show that supernatant protein profiles were different from those of cell pellets of bacteria harvested even in the late growth phase. Furthermore, the secretion rates for UreA and UreB were different.
Significance of Surface Localization of Cytoplasmic Proteins
The ability of released cytoplasmic proteins of H. pylori to bind to the cell surface is considered to be biologically important. For example, the surface locations of catalase and SOD could enable these enzymes to function more effectively as a defense mechanism against phagocytic attack. In the case of urease, one study indicated that free or extracellular urease is irreversibly inhibited at a pH of <4.5. The authors concluded that external urease is ineffective as an acid-protective device at the lower pH values in the stomach (88). However, in another study, bacteria with only cytoplasmic urease showed significantly reduced survival when exposed to acid in the presence of 5 mM urea when compared to the survival of bacteria with both cytoplasmic and extracellular urease (63). This indicates that cytoplasmic urease activity alone is not enough to enable H. pylori to survive in acid. The authors of this study speculate that possibly the association of urease with the outer membrane of H. pylori protects urease from inactivation by acid. Development of mutants in which urease is located strictly within the cytoplasm would be helpful to resolve the discrepancies between the two studies. The role of urease in mediating acid tolerance in H. pylori is discussed in more detail in this volume (chapter 16).
In addition to mediating acid resistance, other roles have been postulated for surface-associated urease that include binding to mucin at acidic pH (50). To test this possibility, the in vitro adherence of an isogenic urease-negative mutant to gastric epithelial cells was compared with that of the parent urease-positive strain. Significant binding was observed for both strains, indicating that urease was not an adhesin (11). At acidic pH in the absence of urea, viability of the organism was reduced and consequently so was binding of the organism to gastric epithelial cells (11, 12).
The observation that urease, HspB, and catalase are surface associated helps explain how they can serve as vaccine components in animal trials (35, 36, 70, 87). However, it is likely that in vivo a subpopulation of H. pylori cells do not carry these antigens on the cell surface, and such a subpopulation may therefore evade the protective effect of the vaccine. In support of this, vaccination studies with urease against H. pylori in monkeys provided only a very low rate of protection (22) or no protection (92) against infection.
Outer Membrane Proteins of H. pylori
Many surface proteins of pathogenic bacteria play important roles in adhesion, colonization, and the immune response. In addition, some outer membrane proteins (OMPs) perform transport functions essential for metabolism, while maintaining the selective permeability of the outer membrane to substances such as antibiotics. Although the production of a large number of cell-envelope proteins in H. pylori has been inferred by genome sequence annotation, functional characterization of most of these has not been reported. Table 1 lists cell-envelope proteins for which biological data are available. Type IV secretion components inferred from analysis of the cag pathogenicity island (15), and the β-barrel domain of the vacuolating cytotoxin (16), have been omitted from this discussion as they are treated elsewhere in this volume (chapters 9 and 19).
Table 1
Cell-envelope proteins of H. pylori.
Porins
Porins of H. pylori were first identified by Hancock, Trust, and coworkers (33) using two-dimensional gel electrophoresis to distinguish heat-modifiable OMPs. Because of the compact structure of the porin β-barrel, monomers are still folded and therefore more mobile than the fully denatured proteins that can be produced by extended boiling in sodium dodecyl sulfate (42). Ultimately five porins were identified, purified, and functionally characterized with this approach (20, 33), and labeled HopA through E.
All five proteins are nonspecific water-filled channels, with minimal selectivity for anions or cations of varying size (20). Conductance in a model membrane system was similar to that of other nonspecific bacterial porins, showing a linear relationship between salt concentration and conductance (33). Compared to other gastrointestinal bacteria such as Campylobacter spp., a relatively small channel size was inferred from the conductivity measurements that is more similar to that of the porins of antibiotic-resistant Pseudomonas spp. (33). This could contribute to the resistance of H. pylori to hydrophilic antibiotics. The functional characterization of H. pylori porins has recently been extended by expression in E. coli and linker mutagenesis (7). The resulting model for the membrane topology of HopE provides a basis for predicting the organization of HopE paralogs, but a systematic public-domain biochemical analysis of these proteins is highly desirable.
H. pylori porins or their paralogs have been implicated in a number of important aspects of host-pathogen interactions. Tufano and colleagues (101) described a range of immunomodulatory responses elicited by a 30-kDa protein from H. pylori, which could be identical to HopE. Since six of six H. pylori-infected patients tested produced antibody against HopE (33), we suggest that some of these effects on the host might be mitigated or abolished by neutralizing antibody. Further studies will be required to substantiate or extend immunomodulatory properties of H. pylori OMPs.
A high degree of sequence identity between the limited HopA–E protein sequences prompted the suggestion that they formed a family of related proteins (33), which was validated by subsequent genome analysis (1, 99). Tomb and coworkers annotated 32 genes belonging to a porin-OMP paralogous gene family, among which the Hops were identified. Further analysis has divided these genes into three paralogous families (1), the largest having 20 and 21 members in strain J99 and strain 26695, respectively (1, 97). These divisions are based on one homologous region at the amino terminal end and seven domains showing similarity at the carboxy-terminal end, the latter including the characteristic alternating hydrophobic residues. Functional characterization of many of these proteins has not been reported, and it is currently unclear how many are porins. Some have other functions related to pathogenesis, such as adhesion (see below).
Adhesins
H. pylori binds to a number of carbohydrate ligands on epithelial cells, including fucosylated glycoproteins such as the Leb antigen (9). The corresponding H. pylori adhesin BabA was purified by affinity tagging, and two babA alleles (babA1 and babA2) were cloned (52). The DNA sequence determination of these alleles and a related gene babB revealed that BabA2 is the expressed Leb adhesin, the babA1 gene being silent because of lack of an initiation codon. The sequences surprisingly showed that they were members of the paralogous gene family of OMPs previously discussed: babA1 and babB correspond to HP1243 and HP896, respectively, whereas babA2 is absent from strain 26695 (52, 99). Some of these genes have been predicted to be phase variable because homopolymeric tracts and dinucleotide repeats present in the signal sequence encoding regions are likely susceptible to slipped strand repair (99). However, when the two published H. pylori genome sequences were compared (1), the predicted expression state of five OMPs was identical in the two strains. The significance of the potential for phase variation in these paralogs is currently unclear.
Another suspected adhesin, the HopZ protein (84), was recently identified in the outer membrane fraction, and the relevant gene was cloned and sequenced from 15 strains. The protein was shown to mediate binding to gastric epithelial cells, but it did not function as a porin. Although a close homolog of hopZ is present in the genome of strain J99 (jhp007, 92% identity in DNA and protein; our analysis), the authors noted that the HP009 gene in strain 26695 was less conserved (88% amino acid identity), and two allelic variants were distinguishable in the 15 strains sequenced (84).
The alpAB gene locus was identified by Haas and colleagues by shuttle mutagenesis; mutation of either of the genes reduced adhesion to human gastric tissue sections (77). The sequences of these two genes, which are organized in an operon, show highest identity to HP0912 and HP0913 (98), encoding HopC and HopB, respectively. The model for the membrane topology of AlpA presented in that study does not refer to the fact that HopC has been experimentally proven to be a porin (33).
Cell-envelope proteins other than porins have been implicated as adhesins. Among the first was Hpa (see below), initially described as a hemagglutinin (30), but now identified as a flagellar sheath lipoprotein with controversial adhesive properties (53, 79). Another 25-kDa OMP was identified that acts in a lectin-like manner with LPS to mediate attachment of H. pylori to laminin, an abundant glycosylated protein in the basement membrane (103). Although this protein has not been identified, it is most likely a member of the OMP family.
Iron-Related OMPs
Worst and colleagues first identified three heme-binding iron-repressible OMPs that were not expressed under normal in vitro growth conditions (108). H. pylori-infected patients develop antibodies that recognize these proteins (109), emphasizing the likelihood that they are essential for survival in the host. The genome contains four genes encoding presumptive OMPs that have significant residue identity to an iron-regulated OMP, FrpB, of Neisseria meningitidis (Table 1). However, the sizes of these proteins are significantly different from those identified by Worst and colleagues (108). The cell envelope is also likely equipped with cytoplasmic membrane iron transport permeases and transport proteins, reviewed elsewhere (68), but they are poorly characterized.
A single protein, NapA, combines several of the biological properties of OMPs already mentioned, including immunomodulation and adhesion. This protein was detected originally in cell-free water extracts of H. pylori that increased surface expression of CD11b/CD18 on neutrophils and increased adhesiveness to endothelial cells (32). NapA (for neutrophil-activating protein) behaved as a 150,000-Da aggregate during chromatographic purification procedures but had a subunit size of 15,000 Da. It was demonstrated that this protein adhered to four glycosphingolipids in a solid-phase assay; a known ganglioside was preferentially bound (98). The surface localization of NapA was independently reported by another group studying H. pylori proteins that bound to mucins (75). A protein, identified as NapA by amino terminal sequencing, was shown to bind to a number of sulfated oligosaccharides and also to the Lex blood group antigen.
Sequence analysis of napA revealed significant identity with bacterioferritins and conservation of a seven-amino-acid motif around the ferroxidase center (31). This inferred intracellular function is at variance but is not necessarily irreconcilable with the adhesive properties and neutrophil stimulation attributed to NapA, applying the previously discussed precedent for surface localization of cytoplasmic proteins. To complicate matters still further, the NapA protein has attracted close scrutiny because it is a major antigen in the human immune response to H. pylori (reported in reference 100). Montecucco and colleagues showed that NapA has a dodecameric quaternary structure (100) and binds up to 40 atoms of iron per monomer. That study did not discuss the surface localization of NapA, though one of the homologs mentioned is an unpublished surface protein of Lactobacillus rhamnosus (accession AAB88605). It would be appropriate to test the biological significance of the reported properties of NapA in experimental infections with a knockout mutant (if viable) or to test strategies to separate and individually neutralize its reported properties.
Flagella
A detailed discussion of the genetics and genomics of H. pylori motility, chemotaxis, and flagella appears elsewhere in this volume (chapter 21). In this section we will focus primarily on structural components of the flagellar apparatus and the general properties associated with this organelle.
The ultrastructure of the H. pylori cell (see chapter 6) is characterized by the presence of unipolar sheathed flagella. Typically there is at least one flagellum, either terminal or subterminal, but there may be up to eight per cell. Aflagellate cells may arise by reversible variation in a homopolymeric sequence re peat in the fliP gene (56). Flagellar dimensions are an average of 4 μm in length, and a more constant 30-nm overall diameter (38, 54). The flagellar filament often terminates in a bulbous, paddle-like structure (Fig. 2). This is composed of the sheath, a relatively uncommon feature in bacterial flagella, which is a membranous layer continuous with the outer membrane (41). Geis and coworkers (38) showed that the protein, phospholipid, and LPS composition of the sheath was broadly similar to that of the outer membrane, although some significant differences in protein and fatty acid composition were noted. More recently, a protein in H. pylori that was first identified as a hemagglutinin (see below) (30), and then shown to be a lipoprotein (79), was reported to be a specific flagellar sheath protein but was not an adhesin (53). The presumed functions of the sheath are acid resistance and masking of flagellar epitopes, which are conserved within the species (1). These epitopes also cross-react with antibodies to Campylobacter jejuni flagellin (76).
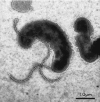
Figure 2
Flagellar configuration of H. pylori. In this negatively stained preparation (1% phosphotungstic acid), terminal and subterminal flagella, hook, sheath, and terminal bulb are visible.
In H. pylori, the filament contains primarily two proteins: a major flagellin of molecular weight 53,000, and a minor component of 54,000 (61, 65, 95). The minor flagellin FlaB was located proximal to the hook, and the major flagellin formed the bulk of the rest of the filament (61). The reliance of the genes for the two flagellins on different sigma factor classes for expression supports a model in which H. pylori varies the ratio of FlaA and FlaB in the flagellum in response to changes in environmental conditions (94). However, both flagellin subunits are necessary for full motility (57). The annotation of the 26695 genome (99) includes two homologs of a presumptive polar flagellin gene flaG of other organisms, but this annotation is not considered robust (81).
The flagellar hook protein (FlgE) of H. pylori (80) is larger than the FlgE proteins of Salmonella enterica serovar Typhimurium or E. coli (67). The assembled hook structure is also larger, which may be related to the physical stresses of terminal flagellar rotation, or motility in the viscous mucosal environment. H. pylori FlgE studies have provided significant insight into flagellar gene regulatory hierarchy (80). Another structural component of the flagellum, the capping protein FliD, has also been genetically characterized (60).
Bacterial cells that have polar rather than peritrichous flagella may have a basal disk, which is thought to dissipate torsion at the cell pole (17, 29). A similar structure may be visualized in H. pylori cells by appropriate negative staining (54, 55). Whether this truly represents a basal disk has not been clarified; the genomic annotation by function transfer from homologs does not help since this structure and its constituent protein are poorly characterized in other bacteria. With regard to the rest of the organelle, there are over 40 flagellar gene homologs in the H. pylori genome that have been reviewed in detail elsewhere (81). The complement of genes suggests that the basal body, hook, and filament are assembled in a manner similar to enterics such as Salmonella, although some major homologs could not be identified: FlgF (proximal rod); FliK (hook length control); FlgD (hook scaffolding); FlgA (P-ring assembly); FliJ (flagellar chaperone). Most of the homologs annotated have not been characterized biochemically. This organism is an attractive model system for elucidating flagellar biogenesis in a small-genome organism.
It has been demonstrated with wild-type strains of varying degrees of motility (25, 26), and with genetically engineered knockout strains (27), that motility is essential for successful colonization in a piglet infection model. Motility is therefore considered a virulence factor for H. pylori (see also chapter 34). One of us (M.C.) has recently shown that strains lacking a flagellum, because of mutation of structural genes, show normal adherence to gastric epithelial cells. In contrast, mutagenesis of the regulatory flagellar gene flbA significantly reduced adherence, suggesting cross-talk with control of production of some other adhesin (13). Mutagenesis of the flagellar export ATPase FliI similarly reduced the production of OMP4 (86), which as yet has no defined function. It seems likely that in this small-genome organism with few transcriptional regulators, overlapping control mechanisms for different export processes are used.
Hemagglutinins
The ability of bacterial cells to agglutinate red blood cells is a convenient assay for detecting bacterial receptors, which usually have affinity for eukaryotic glycolipids or glycoproteins. A confusing array of hemagglutination activities and presumptive hemagglutinins have been reported in the literature, which is briefly summarized here. Further discussion of hemagglutinins and adhesins appears elsewhere in this volume (chapter 34).
The first hemagglutinin described for H. pylori was the HpaA protein (30), which has been discussed above as a constituent of the sheath, and whose adhesion properties are not universally accepted (53, 79). Another neuraminyllactose-sensitive hemagglutinin was isolated by gel filtration and sucrose density gradient ultracentrifugation (47). Further unpublished studies showed that it is a peptidoglycan-associated lipoprotein, which has a neuraminyllactose-binding motif and is conserved in clinical isolates (C. J. Smyth, personal communication). It is possibly identical to the laminin binding protein mentioned above. Another major lipoprotein, Lpp20, was purified from the cell envelope where it was localized to the cytoplasmic membrane and outer membrane (62). Recently Lpp20 has been shown to be protective when used to passively immunize mice, and the surface localization of Lpp20 has been convincingly argued (58). Attempts to isolate hemagglutinins and other adhesins have probably been frustrated by the fact that the activity being purified was due to integral OMPs such as the Hops or BabA. Furthermore, cytoplasmic proteins like DnaK (48) and catalase (83) may become surface localized and meditate binding to glycolipids and glycoproteins, respectively.
Conclusions
Characterization of the cell envelope of H. pylori has identified a number of important features that distinguish it from other bacterial pathogens. These are the simple structure of its peptidoglycan, its unusual cellular fatty acid and lipid profile, molecular mimicry of Lewis antigens by LPS, and the presence of cytoplasmic proteins such as urease and catalase on the cell surface. In addition to these characteristics, polar flagella and a unique repertoire of outer membrane proteins are all likely to contribute to the ability of the organism to colonize the stomach and cause disease. The significance of the outer membrane components, many of which are likely to function as adhesins, is highlighted by the fact that H. pylori devotes a significantly high proportion of its coding capacity to them. Many of the vaccine candidates for H. pylori are proteins found on the cell surface, underlining the importance of further characterization of these proteins and of elucidating the precise mechanism of interaction of the H. pylori cell surface with gastric mucosa.
Acknowledgments
We thank Tadhg Ó Cróinin and Stan Moore for help in preparation of this manuscript and Michael Lane and Doug Hopcroft (HortResearch) for microscopy.
References
- 1.
- Alm R. A., Ling L. S., Moir D. T., King B. L., Brown E. D., Doig P. C., Smith D. R., Noonan B., Guild B. C., de-Jonge B. L., Carmel G., Tummino P. J., Caruso A., Uria-Nickelsen M., Mills D. M., Ives C., Gibson R., Merberg D., Mills S. D., Jiang Q., Taylor D. E., Vovis G. F., Trust T. J. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. [PubMed: 9923682]
- 2.
- Appelmelk B. J., Martin S. L., Monteiro M. A., Clayton C. A., McColm A. A., Zheng P., Verboom T., Maaskant J. J., van den Eijnden D. H., Hokke C. H., Perry M. B., Vandenbroucke-Grauls C. M., Kusters J. G. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in alpha3-fucosyltransferase genes. Infect. Immun. 1999;67:5361–5366. [PMC free article: PMC96892] [PubMed: 10496917]
- 3.
- Appelmelk B. J., Shiberu B., Trinks C., Tapsi N., Zheng P. Y., Verboom T., Maaskant J., Hokke C. H., Schiphorst W. E., Blanchard D., Simoons-Smit I. M., van den Eijnden D. H., Vandenbroucke-Grauls C. M. Phase variation in Helicobacter pylori lipopolysaccharide. Infect. Immun. 1998;66:70–76. [PMC free article: PMC107860] [PubMed: 9423841]
- 4.
- Aspinall G. O., Monteiro M. A. Lipopolysaccharides of Helicobacter pylori strains P466 and MO19: structures of the O antigen and core oligosaccharide regions. Biochemistry. 1996;35:2498–2504. [PubMed: 8652594]
- 5.
- Aspinall G. O., Monteiro M. A., Pang H., Walsh E. J., Moran A. P. Lipopolysaccharide of the Helicobacter pylori type strain NCTC 11637 (ATCC 43504): structure of the O antigen chain and core oligosaccharide regions. Biochemistry. 1996;35:2489–2497. [PubMed: 8652593]
- 6.
- Austin J. W., Doig P., Stewart M., Trust T. J. Structural comparison of urease and a GroEL analog from Helicobacter pylori. J. Bacteriol. 1992;174:7470–7473. [PMC free article: PMC207446] [PubMed: 1358875]
- 7.
- Bina J., Bains M., Hancock R. E. Functional expression in Escherichia coli and membrane topology of porin HopE, a member of a large family of conserved proteins in Helicobacter pylori. J. Bacteriol. 2000;182:2370–2375. [PMC free article: PMC111296] [PubMed: 10762234]
- 8.
- Bode G., Malfertheiner P., Lehnhardt G., Nilius M., Ditschuneit H. Ultrastructural localization of urease of Helicobacter pylori. Med. Microbiol. Immunol. 1993;182:233–242. [PubMed: 8283959]
- 9.
- Borén T., Falk P., Roth K. A., Larson G., Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. [PubMed: 8018146]
- 10.
- Cao P., McClain M. S., Forsyth M. H., Cover T. L. Extracellular release of antigenic proteins by Helicobacter pylori. Infect. Immun. 1998;66:2984–2986. [PMC free article: PMC108299] [PubMed: 9596777]
- 11.
- Clyne M., Drumm B. The urease enzyme of Helicobacter pylori does not function as an adhesin. Infect. Immun. 1996;64:2817–2820. [PMC free article: PMC174146] [PubMed: 8698515]
- 12.
- Clyne M., Labigne A., Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 1995;63:1669–1673. [PMC free article: PMC173208] [PubMed: 7729871]
- 13.
- Clyne M., O'Croinin T., Suerbaum S., Josenhans C., Drumm B. Adherence of isogenic flagella negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect. Immun. 2000;68:4335–4339. [PMC free article: PMC101762] [PubMed: 10858255]
- 14.
- Costa K., Bacher G., Allmaier G., Dominguez-Bello M. G., Engstrand L., Falk P., de Pedro M. A., Garcia-del P. F. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J. Bacteriol. 1999;181:3710–3715. [PMC free article: PMC93848] [PubMed: 10368145]
- 15.
- Covacci A., Rappuoli R. Helicobacter pylori: molecular evolution of a bacterial quasi-species. Curr. Opin. Microbiol. 1998;1:96–102. [PubMed: 10066468]
- 16.
- Cover T. The vacuolating cytotoxin of Helicobacter pylori. Mol. Microbiol. 1996;20:241–246. [PubMed: 8733223]
- 17.
- Curry A., Fox A. J., Jones D. M. A new bacterial flagellar structure found in campylobacters. J. Gen. Microbiol. 1984;130:1307–1310. [PubMed: 6470668]
- 18.
- Cussac V., Ferrero R. L., Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 1992;174:2466–2473. [PMC free article: PMC205883] [PubMed: 1313413]
- 19.
- Doig P., de Jonge B. L., Alm R. A., Brown E. D., Uria-Nickelsen M., Noonan B., Mills S. D., Tummino P., Carmel G., Guild B. C., Moir D. T., Vovis G. F., Trust T. J. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 1999;63:675–707. [PMC free article: PMC103750] [PubMed: 10477312]
- 20.
- Doig P., Exner M. M., Hancock R. E., Trust T. J. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 1995;177:5447–5452. [PMC free article: PMC177350] [PubMed: 7559328]
- 21.
- Doig, P., P. W. O'Toole, and T. J. Trust. 1997. Molecular characterization of H. pylori surface antigens, p. 177–189. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori Protocols. Humana Press Inc., Totowa, N.J. [PubMed: 21351033]
- 22.
- Dubois A., Lee C. K., Fiala N., Kleanthous H., Mehlman P. T., Monath T. Immunization against natural Helicobacter pylori infection in nonhuman primates. Infect. Immun. 1998;66:4340–4346. [PMC free article: PMC108524] [PubMed: 9712786]
- 23.
- Dunn B. E., Roop R. M., Sung C. C., Sharma S. A., Perez-Perez G. I., Blaser M. J. Identification and purification of a cpn60 heat shock protein homolog from Helicobacter pylori. Infect. Immun. 1992;60:1946–1951. [PMC free article: PMC257099] [PubMed: 1563786]
- 24.
- Dunn B. E., Vakil N. B., Schneider B. G., Miller M. M., Zitzer J. B., Peutz T., Phadnis S. H. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect. Immun. 1997;65:1181–1188. [PMC free article: PMC175115] [PubMed: 9119449]
- 25.
- Eaton K., Morgan D. R., Krakowka S. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 1992;37:123–127. [PubMed: 1629897]
- 26.
- Eaton K. A., Morgan D. R., Krakowka S. Campylobacter pylori virulence factors in gnotobiotic piglets. Infect. Immun. 1989;57:1119–1125. [PMC free article: PMC313239] [PubMed: 2925243]
- 27.
- Eaton K. A., Suerbaum S., Josenhans C., Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 1996;64:2445–2448. [PMC free article: PMC174096] [PubMed: 8698465]
- 28.
- Edwards N. J., Monteiro M. A., Faller G., Walsh E. J., Moran A. P., Roberts I. S., High N. J. Lewis X structures in the O antigen side-chain promote adhesion of Helicobacter pylori to the gastric epithelium. Mol. Microbiol. 2000;35:1530–1539. [PubMed: 10760152]
- 29.
- Engelhardt H., Schuster S. C., Baeuerlein E. An archimedian spiral: the basal disk of the Wolinella flagellar motor. Science. 1993;262:1046–1048. [PubMed: 8235620]
- 30.
- Evans D. G., Karjalainen T. K., Evans D. J., Graham D. Y., Lee C. H. Cloning, nucleotide sequence and expression of a gene encoding an adhesin subunit protein of Helicobacter pylori. J. Bacteriol. 1993;175:674–683. [PMC free article: PMC196205] [PubMed: 7678592]
- 31.
- Evans D. J. Jr.,, Evans D. G., Lampert H. C., Nakano H. Identification of four new prokaryotic bacterioferritins, from Helicobacter pylori, Anabaena variabilis, Bacillus subtilis and Treponema pallidum, by analysis of gene sequences. Gene. 1995;153:123–127. [PubMed: 7883175]
- 32.
- Evans D. J. Jr.,, Evans D. G., Takemura T., Nakano H., Lampert H. C., Graham D. Y., Granger D. N., Kvietys P. R. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 1995;63:2213–2220. [PMC free article: PMC173288] [PubMed: 7768601]
- 33.
- Exner M. M., Doig P., Trust T. J., Hancock R. E. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 1995;63:1567–1572. [PMC free article: PMC173190] [PubMed: 7534278]
- 34.
- Ferrero R. L., Cussac V., Courcoux P., Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 1992;174:4212–4217. [PMC free article: PMC206198] [PubMed: 1320607]
- 35.
- Ferrero R. L., Thiberge J. M., Huerre M., Labigne A. Recombinant antigens prepared from the urease subunits of Helicobacter spp: evidence of protection in a mouse model of gastric infection. Infect. Immun. 1994;62:4981–4989. [PMC free article: PMC303216] [PubMed: 7927778]
- 36.
- Ferrero R. L., Thiberge J. M., Kansau I., Wuscher N., Huerre M., Labigne A. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc. Natl. Acad. Sci. USA. 1995;92:6499–6503. [PMC free article: PMC41545] [PubMed: 7604021]
- 37.
- Ge Z., Chan N. W., Palcic M. M., Taylor D. E. Cloning and heterologous expression of an alpha1,3-fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J. Biol. Chem. 1997;272:21357–21363. [PubMed: 9261149]
- 38.
- Geis G., Leying H., Suerbaum S., Mai U., Opferkuch W. Ultrastructure and chemical analysis of Campylobacter pylori flagella. J. Clin. Microbiol. 1989;27:436–441. [PMC free article: PMC267336] [PubMed: 2715319]
- 39.
- Geis G., Leying H., Suerbaum S., Opferkuch W. Unusual fatty acid substitution in lipids and lipopolysaccharides of Helicobacter pylori. J. Clin. Microbiol. 1990;28:930–932. [PMC free article: PMC267839] [PubMed: 2351736]
- 40.
- Goodwin C. S., Chilvers T., Peters M., Collins D., Sly I., McConnell W. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov. respectively. Int. J. Syst. Bacteriol. 1989;39:397–405.
- 41.
- Goodwin C. S., McCulloch R. K., Armstrong J. A., Wee S. H. Unusual cellular fatty acids and distinctive ultrastructure in a new spiral bacterium (Campylobacter pyloridis) from the human gastric mucosa. J. Med. Microbiol. 1985;19:257–267. [PubMed: 3981612]
- 42.
- Hancock, R. E. W., and M. Exner. 1997. Isolation and characterization of porins from H. pylori, p. 191–204. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori Protocols. Humana Press, Totowa, N.J. [PubMed: 21351034]
- 43.
- Haque M., Hirai Y., Yokota K., Mori N., Jahan I., Ito H., Hotta H., Yano I., Kanemasa Y., Oguma K. Lipid profile of Helicobacter spp.: presence of cholesteryl glucoside as a characteristic feature. J. Bacteriol. 1996;178:2065–2070. [PMC free article: PMC177906] [PubMed: 8606185]
- 44.
- Harris A. G., Hazell S. L., Netting A. G. Use of digoxigenin-labelled ampicillin in the identification of penicillin-binding proteins in Helicobacter pylori. J. Antimicrob. Chemother. 2000;45:591–598. [PubMed: 10797079]
- 45.
- Hawtin P. R., Stacey A. R., Newell D. G. Investigation of the structure and localization of the urease of Helicobacter pylori using monoclonal antibodies. J. Gen. Microbiol. 1990;136:1995–2000. [PubMed: 2269872]
- 46.
- Hirai Y., Haque M., Yoshida T., Yokota K., Yasuda T., Oguma K. Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. J. Bacteriol. 1995;177:5327–5333. [PMC free article: PMC177327] [PubMed: 7665522]
- 47.
- Huang J., Keeling P., Smyth C. Identification of erythrocyte-binding antigens in Helicobacter pylori. J. Gen. Microbiol. 1992;138:1503–1513. [PubMed: 1512579]
- 48.
- Huesca M., Goodwin A., Bhagwansingh A., Hoffman P., Lingwood C. A. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 1998;66:4061–4067. [PMC free article: PMC108486] [PubMed: 9712748]
- 49.
- Hunt R. H. Hp and pH—the relevance of gastric acid to the treatment of Helicobacter pylori infection. J. Gastroenterol. 1994;29(Suppl. 7):128–133. [PubMed: 7921145]
- 50.
- Icatlo F. C. Jr.,, Kuroki M., Kobayashi C., Yokoyama H., Ikemori Y., Hashi T., Kodama Y. Affinity purification of Helicobacter pylori urease. Relevance to gastric mucin adherence by urease protein. J. Biol. Chem. 1998;273:18130–18138. [PubMed: 9660771]
- 51.
- Ikeda F., Yokota Y., Mine Y., Tatsuta M. Activity of cefixime against Helicobacter pylori and affinities for the penicillin-binding proteins. Antimicrob. Agents Chemother. 1990;34:2426–2428. [PMC free article: PMC172075] [PubMed: 2088199]
- 52.
- Ilver D., Arnqvist A., Ögren J., Frick I. M., Kersulyte D., Incecik E. T., Berg D. E., Covacci A., Engstrand L., Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. [PubMed: 9430586]
- 53.
- Jones A. C., Logan R. P. H., Foynes S., Cockayne A., Wren B. W., Penn C. W. A flagellar sheath protein of Helicobacter pylori is identical to HpaA, a putative N-acetylneuraminyllactose-binding hemagglutinin, but is not an adhesion for AGS cells. J. Bacteriol. 1997;179:5643–5647. [PMC free article: PMC179448] [PubMed: 9287032]
- 54.
- Jones, D. M., and A. Curry. 1989. The ultrastructure of Campylobacter pylori, p. 48–59. In B. J. Rathbone and R. V. Heatley (ed.), Campylobacter pylori and Gastroduodenal Disease. Blackwell Scientific, Oxford, United Kingdom.
- 55.
- Jones D. M., Curry A., Fox A. J. An ultrastructural study of the gastric campylobacter-like organism `Campylobacter pyloridis'. J. Gen. Microbiol. 1985;131:2335–2341. [PubMed: 4067580]
- 56.
- Josenhans C., Eaton K. A., Thevenot T., Suerbaum S. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 2000;68:4598–4603. [PMC free article: PMC98385] [PubMed: 10899861]
- 57.
- Josenhans C., Labigne A., Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 1995;177:3010–3020. [PMC free article: PMC176987] [PubMed: 7768796]
- 58.
- Keenan J., Oliaro J., Domigan N., Potter H., Aitken G., Allardyce R., Roake J. Immune response to an 18-kilodalton outer membrane antigen identifies lipoprotein 20 as a Helicobacter pylori vaccine candidate. Infect. Immun. 2000;68:3337–3343. [PMC free article: PMC97594] [PubMed: 10816482]
- 59.
- Kidd M., Miu K., Tang L. H., Perez-Perez G. I., Blaser M. J., Sandor A., Modlin I. M. Helicobacter pylori lipopolysaccharide stimulates histamine release and DNA synthesis in rat enterochromaffin-like cells. Gastroenterology. 1997;113:1110–1117. [PubMed: 9322505]
- 60.
- Kim J. S., Chang J. H., Chung S. I., Yum J. S. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 1999;181:6969–6976. [PMC free article: PMC94171] [PubMed: 10559162]
- 61.
- Kostrzynska M., Betts J. D., Austin J. W., Trust T. J. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 1991;173:937–946. [PMC free article: PMC207209] [PubMed: 1704004]
- 62.
- Kostrzynska M., O'Toole P. W., Taylor D. E., Trust T. J. Molecular characterization of a conserved 20 kDa membrane-associated lipoprotein antigen of Helicobacter pylori. J. Bacteriol. 1994;176:5938–5948. [PMC free article: PMC196810] [PubMed: 7928954]
- 63.
- Krishnamurthy P., Parlow M., Zitzer J. B., Vakil N. B., Mobley H. L., Levy M., Phadnis S. H., Dunn B. E. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect. Immun. 1998;66:5060–5066. [PMC free article: PMC108630] [PubMed: 9784504]
- 64.
- Krishnamurthy P., Parlow M. H., Schneider J., Burroughs S., Wickland C., Vakil N. B., Dunn B. E., Phadnis S. H. Identification of a novel penicillin-binding protein from Helicobacter pylori. J. Bacteriol. 1999;181:5107–5110. [PMC free article: PMC94005] [PubMed: 10438788]
- 65.
- Leying H., Suerbaum S., Geis G., Haas R. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 1992;6:2863–2874. [PubMed: 1435261]
- 66.
- Logan S. M., Conlan J. W., Monteiro M. A., Wakarchuk W. W., Altman E. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol. Microbiol. 2000;35:1156–1167. [PubMed: 10712696]
- 67.
- Macnab, R. M. 1996. Flagella and motility, p. 123–145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed. ASM Press, Washington, D.C.
- 68.
- Marais A., Mendz G. L., Hazell S. L., Megraud F. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol. Mol. Biol. Rev. 1999;63:642–674. [PMC free article: PMC103749] [PubMed: 10477311]
- 69.
- McGowan C. C., Necheva A., Thompson S. A., Cover T. L., Blaser M. J. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 1998;30:19–31. [PubMed: 9786182]
- 70.
- Michetti P., Corthesy-Theulaz I., Davin C., Haas R., Vaney A. C., Heitz M., Bille J., Kraehenbuhl J. P., Saraga E., Blum A. L. Immunization of BALB/c mice against Helicobacter felis infection with Helicobacter pylori urease. Gastroenterology. 1994;107:1002–1011. [PubMed: 7926454]
- 71.
- Monteiro M. A., Chan K. H., Rasko D. A., Taylor D. E., Zheng P. Y., Appelmelk B. J., Wirth H. P., Yang M., Blaser M. J., Hynes S. O., Moran A. P., Perry M. B. Simultaneous expression of type 1 and type 2 Lewis blood group antigens by Helicobacter pylori lipopolysaccharides. Molecular mimicry between H. pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. J. Biol. Chem. 1998;273:11533–11543. [PubMed: 9565568]
- 72.
- Moran A. P., Helander I. M., Kosunen T. U. Compositional analysis of Helicobacter pylori rough-form lipopolysaccharides. J. Bacteriol. 1992;174:1370–1377. [PMC free article: PMC206434] [PubMed: 1735724]
- 73.
- Moran A. P., Lindner B., Walsh E. J. Structural characterization of the lipid A component of Helicobacter pylori rough- and smooth-form lipopolysaccharides. J. Bacteriol. 1997;179:6453–6463. [PMC free article: PMC179563] [PubMed: 9335296]
- 74.
- Muotiala A., Helander I. M., Pyhala L., Kosunen T. U., Moran A. P. Low biological activity of Helicobacter pylori lipopolysaccharide. Infect. Immun. 1992;60:1714–1716. [PMC free article: PMC257055] [PubMed: 1548097]
- 75.
- Namavar F., Sparrius M., Veerman E. C., Appelmelk B. J., Vandenbroucke-Grauls C. M. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect. Immun. 1998;66:444–447. [PMC free article: PMC107925] [PubMed: 9453593]
- 76.
- Newell D. G. Identification of the outer membrane proteins of Campylobacter pyloridis and antigenic cross-reactivity between C. pyloridis and C. jejuni. J. Gen. Microbiol. 1987;133:163–170. [PubMed: 3309141]
- 77.
- Odenbreit S., Till M., Hofreuter D., Faller G., Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 1999;31:1537–1548. [PubMed: 10200971]
- 78.
- Odenbreit S., Wieland B., Haas R. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase-deficient mutant strain. J. Bacteriol. 1996;178:6960–6967. [PMC free article: PMC178599] [PubMed: 8955320]
- 79.
- O'Toole P. W., Janzon L., Doig P., Huang J., Kostrzynska M., Trust T. J. The putative neuraminyllactose-binding hemagglutinin HpaA of Helicobacter pylori CCUG 17874 is a lipoprotein. J. Bacteriol. 1995;177:6049–6057. [PMC free article: PMC177441] [PubMed: 7592366]
- 80.
- O'Toole P. W., Kostrzynska M., Trust T. J. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol. Microbiol. 1994;14:691–703. [PubMed: 7891557]
- 81.
- O'Toole P. W., Lane M. C., Porwollik S. Helicobacter pylori motility. Microb. Infect. 2000;2:1207–1214. [PubMed: 11008110]
- 82.
- Page W. J., Taylor D. E. Comparison of methods used to separate the inner and outer membranes of cell envelopes of Campylobacter spp. J. Gen. Microbiol. 1988;134:2925–2932. [PubMed: 2474628]
- 83.
- Pantzar M., Ljungh Å., Wadström T. A vitronectin binding protein of Helicobacter pylori is identical with catalase. Gut. 1999;45(Suppl. III):A27.
- 84.
- Peck B., Ortkamp M., Diehl K. D., Hundt E., Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. [PMC free article: PMC148566] [PubMed: 10454640]
- 85.
- Phadnis S. H., Parlow M. H., Levy M., Ilver D., Caulkins C. M., Connors J. B., Dunn B. E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 1996;64:905–912. [PMC free article: PMC173855] [PubMed: 8641799]
- 86.
- Porwollik S., Noonan B., O'Toole P. W. Molecular characterization of a flagellar export locus of Helicobacter pylori. Infect. Immun. 1999;67:2060–2070. [PMC free article: PMC115938] [PubMed: 10225855]
- 87.
- Radcliff F. J., Hazell S. L., Kolesnikow T., Doidge C., Lee A. Catalase, a novel antigen for Helicobacter pylori vaccination. Infect. Immun. 1997;65:4668–4674. [PMC free article: PMC175669] [PubMed: 9353048]
- 88.
- Scott D. R., Weeks D., Hong C., Postius S., Melchers K., Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. [PubMed: 9428219]
- 89.
- Simoons-Smit I. M., Appelmelk B. J., Verboom T., Negrini R., Penner J. L., Aspinall G. O., Moran A. P., Fei S. F., Shi B. S., Rudnica W., Savio A., de Graaff J. Typing of Helicobacter pylori with monoclonal antibodies against Lewis antigens in lipopolysaccharide. J. Clin. Microbiol. 1996;34:2196–2200. [PMC free article: PMC229216] [PubMed: 8862584]
- 90.
- Smeets L. C., Boomkens S. Y., Bijlsma J. J. E., Vandenbroucke-Grauls C. M., Kusters J. G. Identification of an essential transformation gene in Helicobacter pylori. Gut. 1999;45(Suppl. III):A17.
- 91.
- Smith J. I., Drumm B., Neumann A. W., Policova Z., Sherman P. M. In vitro surface properties of the newly recognized gastric pathogen Helicobacter pylori. Infect. Immun. 1990;58:3056–3060. [PMC free article: PMC313610] [PubMed: 2387633]
- 92.
- Solnick J. V., Canfield D. R., Hansen L. M., Torabian S. Z. Immunization with recombinant Helicobacter pylori urease in specific-pathogen-free rhesus monkeys (Macaca mulatta). Infect. Immun. 2000;68:2560–2565. [PMC free article: PMC97459] [PubMed: 10768944]
- 93.
- Spiegelhalder C., Gerstenecker B., Kersten A., Schiltz E., Kist M. Purification of Helicobacter pylori superoxide dismutase and cloning and sequencing of the gene. Infect. Immun. 1993;61:5315–5325. [PMC free article: PMC281317] [PubMed: 8225605]
- 94.
- Suerbaum S. The complex flagella of gastric Helicobacter species. Trends Microbiol. 1995;3:168–170. [PubMed: 7627454]
- 95.
- Suerbaum S., Josenhans C., Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 1993;175:3278–3288. [PMC free article: PMC204724] [PubMed: 8501031]
- 96.
- Suerbaum S., Thiberge J. M., Kansau I., Ferrero R. L., Labigne A. Helicobacter pylori hspA-hspB heat-shock gene cluster: nucleotide sequence, expression, putative function and immunogenicity. Mol. Microbiol. 1994;14:959–974. [PubMed: 7715457]
- 97.
- Telford J. L., Ghiara P., Dell'Orco M., Comanducci M., Burroni D., Bugnoli M., Tecce M. F., Censini S., Covacci A., Xiang Z. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J. Exp. Med. 1994;179:1653–1658. [PMC free article: PMC2191472] [PubMed: 8163943]
- 98.
- Teneberg S., Miller-Podraza H., Lampert H. C., Evans, Jr D. J., Evans D. G., Danielsson D., Karlsson K. A. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J. Biol. Chem. 1997;272:19067–19071. [PubMed: 9228091]
- 99.
- Tomb J.-F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., Ketchum K. A., Klenk H. P., Gill S., Dougherty B. A., Nelson K., Quackenbush J., Zhou L., Kirkness E. F., Peterson S., Loftus B., Richardson D., Dodson R., Khalak H. G., Glodek A., McKenney K., Fitzegerald L. M., Lee N., Adams M. D., Hickey E., Berg D. E., Gocayne J. D., Utterback T. R., Peterson J. D., Kelley J. M., Cotton M. D., Venter J. C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. [PubMed: 9252185]
- 100.
- Tonello F., Dundon W. G., Satin B., Molinari M., Tognon G., Grandi G., Del Giudice G., Rappuoli R., Montecucco C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 1999;34:238–246. [PubMed: 10564468]
- 101.
- Tufano M. A., Rossano F., Catalanotti P., Liguori G., Capasso C., Ceccarelli M. T., Marinelli P. Immunobiological activities of Helicobacter pylori porins. Infect. Immun. 1994;62:1392–1399. [PMC free article: PMC186292] [PubMed: 8132346]
- 102.
- Vacheron M. J., Guinand M., Francon A., Michel G. Characterisation of a new endopeptidase from sporulating Bacillus sphaericus which is specific for the gamma-d-glutamyl-l-lysine and gamma-d-glutamyl-(l)meso-diaminopimelate linkages of peptidoglycan substrates. Eur. J. Biochem. 1979;100:189–196. [PubMed: 488088]
- 103.
- Valkonen K. H., Wadstrom T., Moran A. P. Identification of the N-acetylneuraminyllactose-specific laminin-binding protein of Helicobacter pylori. Infect. Immun. 1997;65:916–923. [PMC free article: PMC175069] [PubMed: 9038297]
- 104.
- Vanet A., Labigne A. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect. Immun. 1998;66:1023–1027. [PMC free article: PMC108011] [PubMed: 9488391]
- 105.
- Walsh E. J., Moran A. P. Influence of medium composition on the growth and antigen expression of Helicobacter pylori. J. Appl. Microbiol. 1997;83:67–75. [PubMed: 9246772]
- 106.
- Wang G., Boulton P. G., Chan N. W., Palcic M. M., Taylor D. E. Novel Helicobacter pylori alpha1,2-fucosyltransferase, a key enzyme in the synthesis of Lewis antigens. Microbiology. 1999;145:3245–3253. [PubMed: 10589734]
- 107.
- Wirth H. P., Yang M., Karita M., Blaser M. J. Expression of the human cell surface glycoconjugates Lewis x and Lewis y by Helicobacter pylori isolates is related to cagA status. Infect. Immun. 1996;64:4598–4605. [PMC free article: PMC174419] [PubMed: 8890213]
- 108.
- Worst D. J., Otto B. R., de Graaff J. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect. Immun. 1995;63:4161–4165. [PMC free article: PMC173585] [PubMed: 7558334]
- 109.
- Worst D. J., Sparrius M., Kuipers E. J., Kusters J. G., de Graaff J. Human serum antibody response against iron-repressible outer membrane proteins of Helicobacter pylori. FEMS Microbiol. Lett. 1996;144:29–32. [PubMed: 8870248]
- Review Lipopolysaccharide Lewis Antigens.[Helicobacter pylori: Physiolog...]Review Lipopolysaccharide Lewis Antigens.Appelmelk BJ, Vandenbroucke-Grauls CMJE. Helicobacter pylori: Physiology and Genetics. 2001
- Review Helicobacter pylori lipopolysaccharide-mediated gastric and extragastric pathology.[J Physiol Pharmacol. 1999]Review Helicobacter pylori lipopolysaccharide-mediated gastric and extragastric pathology.Moran AP. J Physiol Pharmacol. 1999 Dec; 50(5):787-805.
- Review Pathogenic properties of Helicobacter pylori.[Scand J Gastroenterol Suppl. 1...]Review Pathogenic properties of Helicobacter pylori.Moran AP. Scand J Gastroenterol Suppl. 1996; 215:22-31.
- Review The gastric biology of Helicobacter pylori.[Annu Rev Physiol. 2003]Review The gastric biology of Helicobacter pylori.Sachs G, Weeks DL, Melchers K, Scott DR. Annu Rev Physiol. 2003; 65:349-69. Epub 2002 May 1.
- Role of the alpAB proteins and lipopolysaccharide in adhesion of Helicobacter pylori to human gastric tissue.[Int J Med Microbiol. 2002]Role of the alpAB proteins and lipopolysaccharide in adhesion of Helicobacter pylori to human gastric tissue.Odenbreit S, Faller G, Haas R. Int J Med Microbiol. 2002 Sep; 292(3-4):247-56.
- Cell Envelope - Helicobacter pyloriCell Envelope - Helicobacter pylori
Your browsing activity is empty.
Activity recording is turned off.
See more...
