NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter pylori: Physiology and Genetics. Washington (DC): ASM Press; 2001.
Urease is produced by numerous taxonomically diverse bacterial species, including normal flora and nonpathogens. Also, urease has been demonstrated as a potent virulence factor for some species, including Proteus mirabilis (51), Staphylococcus saprophyticus (36), and Helicobacter pylori (23). Urease is central to H. pylori metabolism and virulence, is necessary for its colonization of the gastric mucosa, and is a potent immunogen that elicits a vigorous immune response. This enzyme is used for taxonomic identification and for diagnosis and follow-up after treatment, and is a vaccine candidate. Urease represents an interesting model for metalloenzyme studies. Before the discovery of H. pylori, humans were thought to produce "gastric urease.'' It is now known that the source of this notable protein is this bacterium, which colonizes the gastric mucosa of humans.
Enzymology
Enzymatic Reaction
Urease (urea amidohydrolase: EC 3.5.1.5) catalyzes the hydrolysis of urea to yield ammonia and carbamate. The latter compound spontaneously decomposes to yield another molecule of ammonia and carbonic acid:
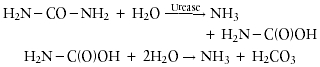
In aqueous solutions, the released carbonic acid and the two molecules of ammonia are in equilibrium with their deprotonated and protonated forms, respectively. The net effect of these reactions is an increase in pH.
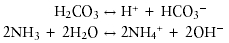
Kinetic Constants and Optima
The kinetic parameters of purified H. pylori urease have been calculated using rates of hydrolysis measured over a range of urea concentrations from 0 to 5 mM at 23°C; the Km ranged from 0.17 to 0.48 mM (21, 27, 46). With purified enzyme under saturating conditions, specific activities ranged from 1,100 to 1,700 μmol of urea/min/mg of protein (21, 46). Although H. pylori has a Km that reflects a higher affinity for substrate than the ureases of other species, this appears to be appropriate to the niche of the bacterium. Assuming that the physiological concentrations of urea to which H. pylori is exposed are the same as those found in serum (1.7 to 3.4 mM), then urease would be saturated and working at its Vmax when the bacterium colonizes the gastric mucosa. Optimal enzyme activity was observed at 43°C for urease in cell lysates (69), and its activity was not inhibited by 0.02% azide (69).
Active Site and Catalysis
The active site of the enzyme is found in the UreB subunit and comprises amino acid residues found throughout the primary structure that are brought into proximity in the tertiary structure (49). Active site residues were identified by examining the results of site-directed mutagenesis (62, 80, 94), studies of apoprotein activation (81), and structural determinations by X-ray crystallography (49, 50). With a numbering specific for H. pylori UreB (55), residues His-136, His-138, Lys-219, His-248, His-274, and Asp-362 come in direct contact with the two nickel ions, urea, or a water molecule within the active site. In addition, His-322 is near the active site and acts as a general base in the catalysis.
The mechanism by which urea is hydrolyzed follows the scheme first described by Zerner's group for the jack bean urease (20). Urea binds in O-coordination to one nickel ion aided by His-221. As an active base, His-322 activates a water molecule bound to the other nickel ion. Attack by the metal-coordinated hydroxide on the substrate carbon atom results in a tetrahedral intermediate that bridges the two nickel sites, a proton is transferred to the intermediate with accompanying ammonia release, and water displaces the carbamate to complete the cycle.
Specific peptide inhibitors of H. pylori urease activity have been identified by screening tetradodecamer and hexamer combinatorial libraries using phage display. The 24-mer TFLPQPRCSALL-RYLSEDGVIVPS and the 6-mer YDFYWW inhibit H. pylori urease with Ki of 47 and 30 μM, respectively, but not the Bacillus pasteurii enzyme (44).
Protein Structure
Urease is a high molecular weight multisubunit metalloenzyme. It appears that all ureases are closely related and have similar mechanisms of catalysis. It is interesting that such a complex protein is required to carry out a simple hydrolysis.
Purification
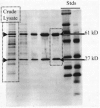
H. pylori synthesizes an extraordinary amount of urease. The purified enzyme, however, is not significantly more active than purified ureases from other species, but it simply represents a larger proportion of total cell protein in this species. When crude lysates are electrophoresed on sodium dodecyl sulfate (SDS)-polyacrylamide gels and stained with Coomassie Blue, the two urease subunits of apparent molecular sizes of 66 and 29.5 kDa appear as extremely prominent bands (46) (Fig. 1). When other urease-positive bacterial species are examined in this manner, urease subunits are not produced in sufficient quantities to detect such bands on the gels.
The native protein has been purified from H. pylori by isolation from the cytosol (46) or by elution from the cell surface with low ionic strength solvents (21, 27, 86, 99). For purifications from the cytosol, French press cell lysates are chromatographed on DEAE-Sepharose, phenyl-Sepharose, Mono-Q, and Sepharose 6 resins. With this scheme, purified urease represents 6% of the soluble protein of crude extract and has an estimated native molecular size of 550 kDa. On the basis of subunit size, a 1:1 subunit ratio measured by scanning densitometry of Coomassie Blue-stained SDS-polyacrylamide gels, and estimated native molecular mass, the data are consistent with a stoichiometry of (29.6-61 kDa)6 for the structure of the native enzyme. The enzyme has an isoelectric point between 5.90 and 5.99 (21, 27, 99).
Appearance in Electron Micrographs
Purified native urease has been examined by transmission electron microscopy and appears as a round, doughnut-shaped, hexagonal particle with a darkly staining core (3, 22, 99). The enzyme is 13 nm in diameter and displays threefold rotational symmetry. Urease eluted from the bacterial cell surface co-purifies with a 60-kDa chaperonin heat shock protein. This heat shock protein displays a very high native molecular weight similar to that of urease and assembles into a macromolecular structure that is also similar in appearance to urease (3, 22, 26).
X-Ray Diffraction Studies
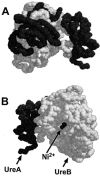
The crystal structure of the related urease from Klebsiella aerogenes has been solved by X-ray diffraction (49, 50). Owing to the high degree of homology between all bacterial ureases, it can be inferred that H. pylori urease shares a similar structure. The K. aerogenes enzyme is only about half the molecular weight of the H. pylori enzyme and contains only three copies of each subunit. Therefore, the H. pylori urease may comprise two of the quaternary structures that are resolved for the Klebsiella enzyme. However, the precise arrangement for the holoenzyme is not known. By using the known crystal structure, a configuration of the H. pylori UreA-UreB heterodimer can be projected (Fig. 2).
Nickel
In common with other ureases, H. pylori contains nickel ions in the active site. The nickel content of H. pylori urease has been measured with atomic absorption spectroscopy (42). All ureases, whether from plants, fungi, or bacteria, that have been analyzed also have nickel ions as a component of the active enzyme (41). On the basis of findings of many studies, two nickel ions (Ni2+) appear to be present in each active site. Because H. pylori has six copies each of the two distinct subunits (UreA and UreB), there are six active sites, and thus 12 nickel ions per fully loaded enzyme molecule.
Effect of pH on Urease
Exposure of whole cells of H. pylori to buffer below a pH of 4 results in loss of intracellular urease activity. In soluble enzyme preparations from lysed cells and supernatants, no urease activity is measurable after incubation at a pH of <5 for 30 min. Exposure of H. pylori cells to a pH of 5 or below inhibits overall protein synthesis, including nascent urease. At a pH of 6 or 7, urease represents 10% of the total cell protein (5).
Genetics
Genetic Organization
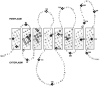
The genes encoding H. pylori urease are located as a single 6.13-kb gene cluster on the chromosome of the bacterium (13, 16, 55) (Fig. 3). Seven contiguous genes, all transcribed in the same direction, are necessary for synthesis of an active enzyme (16, 45, 47). The genes have been designated ureABIEFGH. All of the genes except ureI share homology with urease genes of other species, including Bacillus sp. TB-90 (60), K. aerogenes (56, 73), P. mirabilis (52, 53, 76, 94), Ureaplasma urealyticum (9, 75), Yersinia enterocolitica (17, 92), and the jack bean (85).
Structural Genes
The two structural subunits are encoded by ureA and ureB, the first two genes of the gene cluster (13, 55) (Fig. 3). UreA has a predicted molecular mass of 26.5 kDa, and the predicted masses of UreB are between 60.3 and 61.0 kDa, the sequences differing for various strains. The UreA subunit of Helicobacter species is somewhat unusual because its amino acid sequence is encoded by the single ureA gene, whereas in all other bacterial species it is always encoded by two separate genes. It could be speculated that the two smaller genes of other species may have fused to form H. pylori ureA. Expression of ureA and ureB is sufficient to produce an assembled apoenzyme (45). Under these conditions no nickel ions are inserted into the active site of the enzyme and thus no catalytic activity is present.
Accessory Genes
For synthesis of a catalytically active urease, the accessory genes ureI, ureE, ureF, ureG, and ureH also must be expressed (16) (Fig. 3). Whereas the ureI gene is unique to H. pylori, the remaining accessory genes encode proteins that share homology with gene products of the urease gene clusters of other bacterial species. It is believed that these accessory proteins interact with the apoenzyme and deliver nickel ions to the active site in an energy-dependent process (for a review, see reference 71).
Only few genera, including gastric Helicobacter (88), have the extra ureI gene in the cluster. Weeks et al. demonstrated that UreI is an integral cytoplasmic membrane protein that may form a urea-specific pore (101) (Fig. 3). Importantly, this pore is controlled by external pH via a shift in periplasmic pH (84). There is evidence that the UreI pore opens as the medium pH drops below a pH of 6.5, allowing urea to reach cytoplasmic urease. Thus, as external pH drops, urease activity acts to neutralize the acid; as pH approaches neutrality, substrate is denied to the enzyme and the danger of excessive NH3 is avoided. These authors could not demonstrate UreI-mediated transport in H. pylori itself. However, urea transport in Xenopus oocytes expressing ureI from cRNA appears to be passive (i.e., not driven by proton motive force or ATP hydrolysis), nonsaturable, nonelectrogenic, and temperature-independent. The rate of uptake in oocytes expressing UreI was high at a pH of 5.5 and low at a pH of 7.5. However, Mendz et al. propose that UreI is not involved in urea transport in H. pylori (Mendz et al., personal communication).
Homology to Other Ureases
On the basis of nucleotide sequence of the urease genes of H. pylori and other bacteria, it is certain that all ureases share a common ancestral gene. Despite the fact that ureases are composed of multiple copies of one (jack bean), two (all helicobacters), or three (all other bacterial species) distinct subunits, the amino acid sequences are well conserved. For example, UreA of H. pylori shares 48% and 42% amino acid sequence identity with the corresponding N-terminal sequences of the jack bean urease subunit and the combined sequences of UreA and UreB of P. mirabilis, respectively (53, 55, 85). The entire sequence of the structural subunits of H. pylori shares 58% identity with the urease of K. aerogenes (16, 73). Residues conserved for all ureases and involved in catalysis have been discussed above.
Expression of Catalytically Active Recombinant Urease
A number of requirements were identified to express catalytically active H. pylori urease in E. coli at levels similar to those found in wild-type H. pylori strains (16, 47). Expression of only UreA and UreB was sufficient to produce a normally assembled apoenzyme with no catalytic activity. If accessory genes are coexpressed, a weakly active urease could be produced, but only when Escherichia coli is cultured in minimal salts medium containing no histidine or cysteine (these amino acids chelate free nickel ions). By overexpressing ureA and ureB structural genes in trans to the entire gene cluster, full urease catalytic activity and protein levels could be produced in E. coli, but again only on minimal salts medium lacking histidine and cysteine. Fully active urease, comparable to the wild-type H. pylori enzyme, could be expressed in rich bacteriological medium such as Luria broth, only when NixA (high-affinity Ni2+ transporter) was coexpressed with the entire urease gene cluster and the structural genes were overexpressed (70).
Regulation of Urease Gene Expression
H. pylori does not appear to regulate levels of urease expression by mechanisms such as nitrogen levels, pH, urea induction (71), and iron levels (96), which control urease expression in other species. The ure promoter contains a recognizable – 10 region but not a conserved – 35 region (90). Expression appears to be regulated by selective degradation of urease-encoding mRNA. Akada and colleagues concluded that the gene cluster consists of two operons, ureAB and ureIEFGH, and that primary transcripts of the latter as well as the read-through transcript, ureABIEFGH, are cleaved to produce several mRNA species (1). The ureIEFGH operon may be posttranscriptionally regulated by mRNA decay in response to environmental pH (1).
Other H. pylori genes also may modulate urease gene expression. McGee and colleagues isolated gene bank clones that either stimulated (DNA helicase) or depressed (flbA) urease expression in an E. coli background, providing evidence that flagellar biosynthesis and urease activity are linked (64).
Construction of Mutants
Urease mutants have been constructed by allelic exchange mutagenesis. Antibiotic resistance cassettes were inserted into cloned ureA (McGee and Mobley, unpublished results), ureB, or ureG (28), and these constructs were electroporated into wild-type H. pylori strains. Antibiotic-resistant H. pylori was evaluated for double-crossover mutations in which the wild-type allele was exchanged for the insertionally inactivated allele. Mutants that lacked detectable urease activity were readily selected and had no apparent alteration of growth rates, demonstrating that enzyme activity was not necessary for viability in vitro. These mutants, however, are uniformly avirulent in animal models of infection when tested in pathogenesis studies; ureI mutants are also avirulent (91) (see below). Urease mutants have also been isolated by random insertional mutagenesis using an integration plasmid (8).
Physiology
Substrate Availability
Urea is synthesized in the liver and is found in serum, saliva, and gastric juice at concentrations below 10 mM. It is excreted in urine at high concentrations ranging from 400 to 500 mM (40). A small amount of urea is supplied endogenously to H. pylori by the arginase (RocF)-mediated hydrolysis of l-arginine to l-ornithine (63). Recently, UreI has been postulated to form a urea-specific pore in the cytoplasmic membrane that opens at low pH and closes at high pH, thus regulating urea availability to cytoplasmic urease (101).
Enzyme Localization and Activity of External and Internal Urease
Unlike the ureases of most bacterial species, the enzyme is not strictly cytoplasmic. In aging cultures, urease can be found adherent to the cell surface or shed into the medium (82); this appears to be due to lysis of a subset of the population and readsorption of the protein onto the cell surface of viable bacteria. The importance of the external urease has been debated. Two studies have shown contrasting roles of cytoplasmic versus surface-exposed urease. In the first study, H. pylori was cultured in a way in which the bacteria possessed either cytoplasmic and surface urease or cytoplasmic urease alone; assays were conducted in the presence and absence of flurofamide, a poorly diffusible urease inhibitor (54). Bacteria having only cytoplasmic urease were more susceptible to acid; likewise, E. coli expressing H. pylori urease cytoplasmically was also susceptible to acid. This suggested that surface-exposed urease contributes to resistance to transient acid exposure. In contrast, Scott et al. argued that, because external urease is inactive below a pH of 5 and internal urease has maximal activity below a pH of 5.5, internal urease is most likely responsible for acid tolerance (89).
Use of Ammonia Generated by Urea Hydrolysis
Ammonia, a preferred nitrogen source for bacteria and the product of urea hydrolysis, is assimilated into protein and other nitrogenous compounds in bacteria by a single pathway (83). Glutamine synthetase (EC 6.3.1.2) catalyzes the reaction:
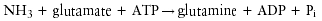
Glutamine, in turn, serves as nitrogen donor for other nitrogenous compounds including alanine, glycine, serine, histidine, tryptophan, CTP, AMP, carbamoyl-phosphate, and glucosamine 6-phosphate.
The 1,443-bp glutamine synthetase gene, glnA, encodes a predicted polypeptide of 481 amino acid residues with a molecular weight of 54,317 (35). In most genera this enzyme activity is regulated posttranslationally by adenylation of the protein. The adenylation site found in most bacterial homologs has consensus sequence NLYDLP, which is replaced in H. pylori by NLFKLT (residues 405 to 410). Since the Tyr (Y) residue is the target of adenylation, and H. pylori glutamine synthetase lacks that residue in four strains examined, no adenylation occurs within this motif and the enzyme is not regulated in this manner. It was not possible to isolate glutamine synthetase-deficient mutants constructed by allelic exchange, suggesting that glutamine synthetase is essential for viability and critical for nitrogen assimilation in H. pylori (35). The enzyme appears active under all physiologic conditions, consistent with the singular niche that the organism occupies.
NixA and Nickel Transport
All ureases contain two nickel ions in each of their active sites; H. pylori urease appears to have six active sites and, thus, 12 nickel ions. To overcome the nickel limitation that probably occurs in the host, H. pylori has developed a high-affinity system to acquire nickel ions. It appears that these ions can be transported into H. pylori by at least two mechanisms. The first is NixA, a cytoplasmic membrane-bound protein of 36,991 molecular weight, which transports nickel ions with a KT of 11.3 nM (70). E. coli expressing NixA transported nickel ions with a Vmax of 1,750 pmol/min/108 bacteria. Topology studies using 21 phoA and 21 lacZ translational fusions revealed that the protein has eight transmembrane domains and a large periplasmic loop with both the N terminus and C terminus residing in the cytosol (Fig. 4) (34). Negatively charged Asp and Glu residues and His residues, which are located in the transmembrane domains, were shown to be critical for active transport of nickel ions (33). Two domains have been identified in NixA and the small family of high-affinity nickel transport proteins as essential for transport: GX2HAXDADH in helix II and GX2FX2GHSSVV in helix III (33).
Insertional inactivation of nixA in H. pylori ATCC 43504 resulted in a 69% decrease in the rate of nickel transport and a 42% reduction in urease activity relative to the parent strain (6). These rates varied among strains but are reduced significantly in the nixA mutants. The fact that nickel transport or urease activity is not totally abolished in these mutants supports the presence of a second mechanism of nickel transport that has yet to be elucidated.
When a nixA mutant of H. pylori SS1 and the parent strain were mixed and used orogastrically to cochallenge mice, the nixA mutant was never recovered by culture of gastric tissue (77). That is, the wild-type always outcompeted the nixA mutant. Interestingly, however, the nixA mutant was able to colonize the gastric mucosa of mice when inoculated alone (i.e., in the absence of the wild type), and the CFU per gram of stomach approached levels achieved in mice infected with the parental strain. These studies indicated that NixA-mediated nickel transport provides a selective advantage for H. pylori in the gastric mucosa.
Metal-Binding Proteins That May Affect Urease Activity
Nickel ions are required for catalytic activity of urease, yet a nixA mutant of H. pylori still retains some urease activity (6). It was concluded from this observation that additional proteins may play a role in nickel binding or transport. Three H. pylori proteins have been identified with predicted nickel-binding or general metal-binding properties.
Hpn is a small protein with a molecular size of 7.1 kDa and 60 amino acid residues (37). Nearly half (47%) of its residues are histidyls, making this protein a strong binder of nickel and zinc ions. Although it is possible to hypothesize that Hpn would serve as a sink for nickel and be used to activate urease, a mutant in which hpn was insertionally inactivated showed no reduction of urease activity following culture in vitro (37). The effects of this mutation have not been examined in vivo, and under these conditions Hpn may have an effect on urease activity.
CadA, a P-type ATPase, has been identified in H. pylori (66). From the gene sequence a 686-amino acid protein is predicted that contains consensus sites for phosphorylation, ATP-binding, and, at the N terminus, segments rich in histidine, methionine, glutamate, and aspartate residues, which may act as a nickel-binding domain. This protein has been shown to be a heavy metal ion (Cd2+, Zn2+, Co2+) exporter that uses ATP as an energy source for transport. Mutation of the gene encoding the ATPase appears to diminish but not abolish (as originally reported) urease activity. These observations suggest that CadA contributes to high levels of urease activity by exporting divalent metal cations that may interfere with Ni2+-metalloenzyme formation (43).
The HspA (heat shock protein A) of H. pylori is 118 amino acid residues long and a homolog of GroES-like heat shock proteins. It is encoded by the first gene, hspA, of a bicistronic cluster (95). Although the predicted protein is highly conserved with respect to its homologs, the last 25 residues of the C terminus include 4 cysteinyls and 6 histidyls, a potential metal-binding motif not found in any of the homologs. Mutation of hspA appears to be lethal for H. pylori and thus the role of this heat shock protein in urease activity cannot be tested directly in the native organism. Expression in E. coli of hspA in trans to the H. pylori urease gene cluster results in slight elevation of urease activity, suggesting that HspA may affect urease activity in wild-type H. pylori.
Pathogenesis
Requirement for Colonization
Although urease is not required for in vitro viability of H. pylori, it is clear that the enzyme is a critical virulence determinant necessary for colonization of the gastric mucosa. A urease-negative mutant, isolated after mutagenesis with nitrosoquanidine of an H. pylori strain, was used to inoculate gnotobiotic piglets. The mutant, which retained only 0.4% of the urease activity of the parent strain, did not colonize any of 10 orally challenged piglets as assessed at 3 and 21 days after challenge, and no pathology was observed in these piglets (23). The parent strain, on the other hand, successfully colonized all seven piglets and caused gastritis. This work was carried further by using an isogenic pair of strains that included the parent strain and an allelic exchange mutant in which ureG had been interrupted with a kanamycin resistance cassette. In these studies the piglets were treated with omeprazole, a proton pump inhibitor, which abolished acid secretion and yielded a neutral pH in their stomach. The parent strain colonized normally in numbers with a mean log10 CFU between 4.4 and 6.9. The urease-negative mutant was unable to colonize the gastric mucosa at normal physiological pH and was recovered only in low numbers (mean log10 CFU, <2) from omeprazole-treated, achlorhydric piglets (24). These results confirmed that urease enzymatic activity was essential for colonization. Importantly, it also implied that the role of urease extended beyond that of just acid protection. Other studies demonstrated the inability of H. pylori urease mutants to colonize the gastric mucosa of nude mice (98) and cynomolgus monkeys (97).
Detection of H. pylori using Urease
Urease Biopsy Test
The high level of expression of urease by H. pylori can be employed for simple detection of the bacterium in gastric biopsies. Samples obtained by endoscopy are placed in a gel containing urea and phenol red (a pH-indicating dye); if H. pylori is present, preformed urease will hydrolyze the urea, raise the pH, and change the color of the phenol red from yellow to red (61, 65). This concept was used in the CLO-test, the first commercially available test for detecting the presence of H. pylori (61). The utility of this test has been repeatedly validated in the literature. Recently, a urease sensor has been developed for endoscopy based on the differential output of two pH-sensitive transistors at the tip of the endoscope. It carries a urea solution that would be hydrolyzed by urease produced by colonizing bacteria (87).
Urease-Positive Colonies after Culture
Freshly cultured H. pylori is strongly urease positive. After 3 to 5 days of culture, pinpoint colonies can be observed and tested for urease activity. H. pylori cultured directly from endoscopic biopsy specimens on serum-based medium virtually always gives a strong urease reaction, which, together with the presence of positive oxidase and catalase reactions, is diagnostic for this species.
Urea Breath Test
Although the urease biopsy test reaction is simple, it requires obtaining biopsies by endoscopy, an invasive procedure. The urea breath test has been developed as a noninvasive procedure that serves as a sensitive and specific, although qualitative, indicator of infection. The patient is given an oral dose of labeled urea, either [13C]urea (39) or [14C]urea (7); if the organism is present, urea will be hydrolyzed and 13CO2 or 14CO2 will be liberated. The labeled carbon dioxide will enter the bloodstream, exchange in the lungs, and be exhaled. The exhaled CO2 is trapped and quantitated in a mass spectrometer for 13CO2 or a scintillation counter for 14CO2. A number of members of the normal anaerobic gut flora are urease positive and potentially could interfere with this test, but data collected thus far indicate that false-positive reactions are rare.
PCR Identification
PCR amplification of H. pylori urease genes has been used in methods to establish the presence of viable or nonviable H. pylori. In one method boiled supernatants from H. pylori strains are used as template for the reaction, and amplification of a 411-bp fragment from ureA is used to identify specifically the bacterium (10, 11, 30, 100). The primers are sensitive and specific for H. pylori and do not react with other Helicobacter spp. In another method a pair of PCR primers, one of which is degenerate, is employed to amplify a 365-bp segment from an area adjacent to the 5′ end of the ureA gene (102). Consistent amplification from gastric juice was reported using this system. PCR amplification from regions of the ureB gene has also been used to detect H. pylori in paraffin-embedded biopsy samples (102).
Typing Systems Based on Urease Genes
There is a significant variation between the DNA sequences of different H. pylori strains. The result is that for a given DNA segment there is heterogeneity between strains regarding sequences and restriction sites. This allowed the development of various typing methods based on urease genes. The first system developed was based on PCR amplification of urease genes (31, 32). PCR amplifications of the structural subunit genes ureA and ureB from clinical isolates yield 2.4-kb PCR products, which digested with HaeIII produce distinct patterns on agarose gels. These patterns allow easy differentiation between strains. In another method to distinguish between strains of H. pylori the ureC gene (no longer considered a urease gene) is amplified, followed by direct DNA sequencing of the PCR product (15). Numerous base-pair changes are detected in ureC, and strains are easily differentiated. Seven subsequent reports confirmed that the urease genes ureA and ureB and the former ureC, subjected to PCR amplification and digestion with restriction endonucleases, can be used to differentiate strains on the basis of patterns on agarose gels, identify the presence of multiple strains in a single biopsy specimen, check for reinfection with the same strain following eradication therapy, and identify similar strains among family members (4, 12, 18, 48, 59, 72, 78). This general strategy has been successfully applied to other H. pylori genes.
Urease in Vaccines
Because infected humans mount a significant immunoglobulin response to urease, it was reasoned that immunization with urease may protect against acquisition of infection. A matter to be elucidated was the form of the protein that should be used in vaccination. Possible candidates included peptides derived from the predicted sequence, isolated subunits (UreA or UreB), apoenzyme, or holoenzyme; but thus far, peptides have not been tested. Two groups of researchers used similar strategies, with the mouse model and an H. felis challenge as a surrogate for H. pylori. Michetti et al. found moderate but nevertheless significant protection when purified urease was used for immunization (67). However, when the urease structural subunits UreA and UreB, purified separately as histidine-tagged fusion proteins on a Ni2+ nitrilotriacetic acid column, were used for immunization, mice were protected from colonization of the gastric mucosa by H. felis. UreB, which harbors the enzyme active site, gave earlier and more complete protection from the challenge strain. In later studies, oral immunization with UreB was used therapeutically for eradication (14). In independent efforts, Ferrero et al. (29) used H. pylori and H. felis UreA and UreB, purified as maltose-binding protein translational fusions, to immunize mice with cholera toxin as the adjuvant. UreB provided more significant protection than UreA, and the H. felis-derived subunits conferred better protection than the H. pylori urease subunits.
Using the holoenzyme would have the advantage of retaining conformational epitopes, but it would be undesirable to retain ureolytic activity in preparations that would be administered by any route because of the ubiquitous presence of urea in humans. Catalytically inactive but fully assembled H. pylori urease was viewed as more desirable and was adopted as a vaccine candidate on the basis of observations of Hu et al. (45). They found that the H. pylori urease apoenzyme, lacking nickel ions in the active site and thus lacking enzymatic activity, could be isolated from E. coli expressing only the ureA and ureB structural subunit genes. These clones lacked the accessory genes necessary for nickel ion incorporation. The apoenzyme was purified according to the precise scheme used for the first reported isolation of the native enzyme, and it eluted from the four resins used for purification (DEAE-Sepharose, Phenyl-Sepharose, MonoQ, and Superose 6) at the same fractions as those observed for the native enzyme (45). This result suggested that the apoenzyme possessed the identical charge, hydrophobicity, shape, and size of the native enzyme and thus was fully assembled lacking only the 12 nickel ions from the active site. On the basis of these data the apoenzyme was adopted as a vaccine candidate.
Recombinant apourease encoded by clones carrying only ureAB has been used for immunization and also shown to provide significant protection for mice against H. felis challenge (58, 74, 79). In these studies, protection was correlated with a high secretory immunoglobulin A titer; in other work, antibody response was found not to be required for protection (25). The results of these investigations provided strong evidence that oral immunization with urease in the presence of adjuvant is a feasible strategy for development of a vaccine for prevention of H. pylori infection in humans. Studies in monkeys with purified urease apoenzyme (93) or in humans using salmonella phoP/phoQ deletion mutant (38) expressing the apoenzyme demonstrated little or no protection against H. pylori infection (2, 19, 68). Other studies in monkeys, however, showed significant reduction in colonization (57).
Acknowledgment
This work was supported in part by Public Health Service Grant AI25567 from the National Institutes of Health.
References
- 1.
- Akada J. K., Shirae M., Takeuchi H., Tsuda M., Nakazawa T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 2000;36:1071–1084. [PubMed: 10844692]
- 2.
- Angelakopoulos H., Hohmann E. L. Pilot study of phoP/phoQ7-deleted Salmonella enterica serovar Typhimurium expressing Helicobacter pylori urease in adult volunteers. Infect. Immun. 2000;68:2135–2141. [PMC free article: PMC97395] [PubMed: 10722611]
- 3.
- Austin J. W., Doig P., Stewart M., Trust T. J. Macromolecular structure and aggregation states of Helicobacter pylori urease. J. Bacteriol. 1991;173:5663–5667. [PMC free article: PMC208295] [PubMed: 1885543]
- 4.
- Bamford K. B., Bickley J., Collins J. S., Johnston B. T., Potts S., Boston V., Owen R. J., Sloan J. M. Helicobacter pylori: comparison of DNA fingerprints provides evidence for intrafamilial infection. Gut. 1993;34:1348–1350. [PMC free article: PMC1374539] [PubMed: 8244100]
- 5.
- Bauerfeind P., Garner R., Dunn B. E., Mobley H. L. T. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. [PMC free article: PMC1027003] [PubMed: 9155571]
- 6.
- Bauerfeind P., Garner R. M., Mobley H. L. T. Allelic exchange mutagenesis of nixa in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 1996;64:2877–2880. [PMC free article: PMC174160] [PubMed: 8698529]
- 7.
- Bell G. D., Weil J., Harrison G., Morden A., Jones P. H., Gant P. N., Trowell J. E., Yoong A. K., Daneshmend T. K., Logan R. F. 14C-urea breath analysis, a non-invasive test for Campylobacter pylori in the stomach. Lancet. 1987;i:1367–1368. [PubMed: 2884468]
- 8.
- Bijlsma J. J., Vandenbroucke-Grauls C. M., Phadnis S. H., Kusters J. G. Identification of virulence genes of Helicobacter pylori by random insertion mutagenesis. Infect. Immun. 1999;67:2433–2440. [PMC free article: PMC115989] [PubMed: 10225906]
- 9.
- Blanchard A. Ureaplasma urealyticum urease genes; use of a UGA tryptophan codon. Mol. Microbiol. 1990;4:669–678. [PubMed: 2191184]
- 10.
- Clayton C. L., Kleanthous H., Coates P. J., Morgan D. D., Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J. Clin. Microbiol. 1992;30:192–200. [PMC free article: PMC265019] [PubMed: 1734052]
- 11.
- Clayton C. L., Kleanthous H., Morgan D. D., Puckey L., Tabaqchali S. Rapid fingerprinting of Helicobacter pylori by polymerase chain reaction and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 1993;31:1420–1425. [PMC free article: PMC265554] [PubMed: 8100240]
- 12.
- Clayton C., Kleanthous H., Tabaqchali S. Detection and identification of Helicobacter pylori by the polymerase chain reaction. J. Clin. Pathol. 1991;44:515–516. [PMC free article: PMC496836] [PubMed: 2066432]
- 13.
- Clayton C. L., Pallen M. J., Kleanthous H., Wren B. W., Tabaqchali S. Nucleotide sequence of two genes from Helicobacter pylori encoding for urease subunits. Nucleic Acids Res. 1990;18:362. [PMC free article: PMC330277] [PubMed: 2326167]
- 14.
- Corthesy-Theulaz I., Porta N., Glauser M., Saraga E., Vaney A.-C., Haas R., Kraehenbuhl J.-P., Blum A. L., Michetti P. Oral immunization with Helicobacter pylori urease B as a treatment against Helicobacter infection. Gastroenterology. 1995;109:115–121. [PubMed: 7797009]
- 15.
- Courcoux P., Freuland C., Piemout Y., Fauchere J. L., Labigne A. Polymerase chain reaction and direct DNA sequencing as a method for distinguishing between different strains of Helicobacter pylori. Rev. Esp. Enf. Dig. 1990;78(Suppl. 1):29–30.
- 16.
- Cussac V., Ferrero R. L., Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 1992;174:2466–2473. [PMC free article: PMC205883] [PubMed: 1313413]
- 17.
- de Koning-Ward T. F., Ward A. C., Robins-Browne R. M. Characterization of the urease-encoding gene complex of Yersinia enterocolitica. Gene. 1994;145:25–32. [PubMed: 8045421]
- 18.
- Desai M., Linton D., Owen R. J., Stanley J. Molecular typing of Helicobacter pylori isolates from asymptomatic, ulcer and gastritis patients by urease gene polymorphism. Epidemiol. Infect. 1994;112:151–160. [PMC free article: PMC2271484] [PubMed: 7907028]
- 19.
- DiPetrillo M. D., Tibbetts T., Kleanthous H., Killeen K. P., Hohmann E. L. Safety and immunogenicity of phoP/phoQ-deleted Salmonella typhi expressing Helicobacter pylori urease in adult volunteers. Vaccine. 1999;18:449–459. [PubMed: 10519934]
- 20.
- Dixon N. E., Riddles P. W., Gazzola C., Blakeley R. L., Zerner B. Jack bean urease (EC 3.5.1.5). V. On the mechanism of action of urease on urea, formamide, acetamide, N-methylurea, and related compounds. Can. J. Biochem. 1980;58:1335–1344. [PubMed: 6788353]
- 21.
- Dunn B. E., Campbell G. P., Perez-Perez G. I., Blaser M. J. Purification and characterization of urease from Helicobacter pylori. J. Biol. Chem. 1990;265:9464–9469. [PubMed: 2188975]
- 22.
- Dunn B. E., Roop R. M., 2nd, Sung C.-C., Sharma S. A., Perez-Perez G. I., Blaser M. J. Identification and purification of a cpn60 heat shock protein homolog from Helicobacter pylori. Infect. Immun. 1992;60:1946–1951. [PMC free article: PMC257099] [PubMed: 1563786]
- 23.
- Eaton K. A., Brooks C. L., Morgan D. R., Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 1991;59:2470–2475. [PMC free article: PMC258033] [PubMed: 2050411]
- 24.
- Eaton K. A., Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 1994;62:3604–3607. [PMC free article: PMC303008] [PubMed: 8063376]
- 25.
- Ermak T. H., Giannasca P. J., Nichols R., Myers G. A., Nedrud J., Weltzin R., Lee C. K., Kleanthous H., Monath T. P. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC cless II-restricted responses. J. Exp. Med. 1998;188:2277–2288. [PMC free article: PMC2212427] [PubMed: 9858514]
- 26.
- Evans D. J. Jr.,, Evans D. G., Engstrand L., Graham D. Y. Urease-associated heat shock protein of Helicobacter pylori. Infect. Immun. 1992;60:2125–2127. [PMC free article: PMC257126] [PubMed: 1348725]
- 27.
- Evans D. J. Jr.,, Evans D. G., Kirkpatrick S. S., Graham D. S. Characterization of the Helicobacter pylori urease and purification of its subunits. Microb. Pathog. 1991;10:15–26. [PubMed: 1857197]
- 28.
- Ferrero R. L., Cussac V., Courcoux P., Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 1992;174:4212–4217. [PMC free article: PMC206198] [PubMed: 1320607]
- 29.
- Ferrero R. L., Thiberge J.-M., Huerre M., Labigne A. Recombinant antigens prepared from the urease subunits of Helicobacter spp.: evidence of protection in a mouse model of gastric infection. Infect. Immun. 1994;62:4981–4989. [PMC free article: PMC303216] [PubMed: 7927778]
- 30.
- Foxall P. A., Hu L.-T., Mobley H. L. T. Amplification of the complete urease structural genes from Helicobacter pylori clinical isolates and cosmid gene bank clones. Rev. Esp. Enf. Dig. 1990;78(Suppl. 1):128–129.
- 31.
- Foxall P. A., Hu L.-T., Mobley H. L. T. Use of polymerase chain reaction-amplified Helicobacter pylori urease structural genes for differentiation of isolates. J. Clin. Microbiol. 1992;30:739–741. [PMC free article: PMC265146] [PubMed: 1313051]
- 32.
- Foxall, P. A., L.-T. Hu, and H. L. T. Mobley. 1991. Detection of Helicobacter pylori urease structural genes by PCR amplification, abstr. 878. Abst. 91st Annual Meet. Am. Soc. Microbiol., 1991. American Society for Microbiology, Washington, D.C.
- 33.
- Fulkerson J. F. Jr.,, Garner R. M., Mobley H. L. T. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J. Biol. Chem. 1998;273:235–241. [PubMed: 9417070]
- 34.
- Fulkerson J. F. Jr, Mobley H. L. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III. J. Bacteriol. 2000;182:1722–1730. [PMC free article: PMC94471] [PubMed: 10692379]
- 35.
- Garner R. G., Fulkerson J. F. Jr, Mobley H. L. T. Helicobacter pylori glutamine synthetase lacks features associated with transcriptional and posttranslational regulation. Infect. Immun. 1998;66:1839–1847. [PMC free article: PMC108133] [PubMed: 9573059]
- 36.
- Gatermann S., Marre R. Cloning and expression of Staphylococcus saprophyticus urease gene sequences in Staphylococcus carnosus and contribution of the enzyme to virulence. Infect. Immun. 1989;57:2998–3002. [PMC free article: PMC260761] [PubMed: 2777370]
- 37.
- Gilbert J. V., Ramakrishna J., Sunderman F. W. Jr, Wright A., Plaut A. G. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect. Immun. 1995;63:2682–2688. [PMC free article: PMC173359] [PubMed: 7790085]
- 38.
- Gomez-Duarte O. G., Bumann D., Meyer T. F. The attenuated Salmonella vaccine approach for the control of Helicobacter pylori-related diseases. Vaccine. 1999;17:1667–1673. [PubMed: 10194821]
- 39.
- Graham D. Y., Klein P. D., Evans D. J., Alpert L. C., Opekun A. R., Boutton T. W. Campylobacter pyloridis detected by the 13C-urea test. Lancet. 1987;i:1174–1177. [PubMed: 2883491]
- 40.
- Griffith D. P., Musher D. M., Itin C. Urease: the primary cause of infection-induced urinary stones. Invest. Urol. 1976;13:346–350. [PubMed: 815197]
- 41.
- Hausinger R. P. Nickel utilization by microorganisms. Microbiol. Rev. 1987;51:22–24. [PMC free article: PMC373090] [PubMed: 3104749]
- 42.
- Hawtin P. R., Delves H. T., Newell D. G. The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol. Lett. 1991;77:51–54. [PubMed: 2004697]
- 43.
- Herrmann L., Schwan D., Garner R., Mobley H. L., Haas R., Schafer K. P., Melchers K. Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol. Microbiol. 1999;33:524–536. [PubMed: 10417643]
- 44.
- Houimel M., Mach J. P., Corthesy-Theulaz I., Corthesy B., Fisch I. New inhibitors of Helicobacter pylori urease holoenzyme selected from phage-displayed peptide libraries. Eur. J. Biochem. 1999;262:774–780. [PubMed: 10411639]
- 45.
- Hu L.-T., Foxall P. A., Russell R., Mobley H. L. T. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Infect. Immun. 1992;60:2657–2666. [PMC free article: PMC257217] [PubMed: 1612735]
- 46.
- Hu L.-T., Mobley H. L. T. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 1990;58:992–998. [PMC free article: PMC258572] [PubMed: 2318539]
- 47.
- Hu L.-T., Mobley H. L. T. Expression of catalytically active recombinant Helicobacter pylori urease at wild type levels in Escherichia coli. Infect. Immun. 1993;61:2563–2569. [PMC free article: PMC280885] [PubMed: 8500893]
- 48.
- Hurtado A., Owen R. J. Identification of mixed genotypes in Helicobacter pylori from gastric biopsy tissue by analysis of urease gene polymorphisms. FEMS Immun. Med. Microbiol. 1994;8:307–313. [PubMed: 7914791]
- 49.
- Jabri E., Carr M. B., Hausinger R. P., Karplus P. A. The crystal structure of urease from Klebsiella aerogenes at 2 Å resolution. Science. 1995;268:998–1004. [PubMed: 7754395]
- 50.
- Jabri E., Lee M. H., Hausinger R. P., Karplus P. A. Preliminary crystallographic studies of ureases from jack bean and from Klebsiella aerogenes. J. Mol. Biol. 1992;227:934–937. [PubMed: 1404395]
- 51.
- Jones B. D., Lockatell C. V., Johnson D. E., Warren J. W., Mobley H. L. T. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 1990;58:1120–1123. [PMC free article: PMC258592] [PubMed: 2180821]
- 52.
- Jones B. D., Mobley H. L. T. Proteus mirabilis urease: genetic organization, regulation, and expression of structural genes. J. Bacteriol. 1988;170:3342–3349. [PMC free article: PMC211300] [PubMed: 2841283]
- 53.
- Jones B. D., Mobley H. L. T. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J. Bacteriol. 1989;171:6414–6422. [PMC free article: PMC210529] [PubMed: 2687233]
- 54.
- Krishnamurthy P., Parlow M., Zitzer J. B., Vakil N. B., Mobley H. L. T., Levy M., Phadnis S. H., Dunn B. E. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect. Immun. 1998;66:5060–5066. [PMC free article: PMC108630] [PubMed: 9784504]
- 55.
- Labigne A. V. Cussac, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J. Bacteriol. 1991;173:1920–1931. [PMC free article: PMC207722] [PubMed: 2001995]
- 56.
- Lee M. H., Mulrooney S. B., Renner M. J., Markowicz Y., Hausinger R. P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, ureG) are involved in nickel metallocenter biosynthesis. J. Bacteriol. 1992;174:4324–4330. [PMC free article: PMC206216] [PubMed: 1624427]
- 57.
- Lee C. K., Soike K., Hill J., Georgakopoulos K., Tibbitts T., Ingrassia J., Gray H., Boden J., Kleanthous H., Giannasca P., Ermak T., Weltzin R., Banchard J., Monath T. P. Immunization with recombinant Helicobacter pylori urease decreases colonization levels following experimental infection of rhesus monkeys. Vaccine. 1999;17:1493–1505. [PubMed: 10195786]
- 58.
- Lee C. K., Weltzin R., Thomas W. D. Jr, Kleanthous H., Ermak T. H., Soman G., Hill J. E., Ackerman S. K., Monath T. P. Oral immunization with recombinant Helicobacter pylori urease induces secretory IgA antibodies and protects mice from challenge with Helicobacter felis. J. Infect. Dis. 1995;172:161–172. [PubMed: 7797906]
- 59.
- Lopez C. R., Owen R. J., Desai M. Differentiation between isolates of Helicobacter pylori by PCR-RFLP analysis of urease A and B genes and comparison with ribosomal RNA gene patterns. FEMS Microbiol. Lett. 1993;110:37–44. [PubMed: 8100546]
- 60.
- Maeda M., Hidaka M., Nakamura A., Masaki H., Uozumi T. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J. Bacteriol. 1994;176:432–42. [PMC free article: PMC205067] [PubMed: 8288539]
- 61.
- Marshall B. J., Warren J. R., Francis G. J., Langton S. R., Goodwin C. S., Blincow E. D. Rapid urease test in the management of Campylobacter pyloridis-associated gastritis. Am. J. Gastroenterol. 1987;82:200–210. [PubMed: 3548326]
- 62.
- Martin P. R., Hausinger R. P. Site-directed mutagenesis of the active site cysteine in Klebsiella aerogenes urease. J. Biol. Chem. 1992;267:20024–20027. [PubMed: 1400317]
- 63.
- McGee D. J., Radcliff F. J., Mendz G. L., Ferrero R. L., Mobley H. L. T. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 1999;181:7314–7322. [PMC free article: PMC103695] [PubMed: 10572136]
- 64.
- McGee D. J., May C. A., Garner R. M., Himpsl J. M., Mobley H. L. T. Isolation of Helicobacter pylori genes that modulate urease activity. J. Bacteriol. 1999;181:2477–2484. [PMC free article: PMC93674] [PubMed: 10198012]
- 65.
- McNulty C. A. M., Wise R. Rapid diagnosis of Campylobacter-associated gastritis. Lancet. 1985;i:1443. [PubMed: 2861379]
- 66.
- Melchers K., Weitzenegger T., Buhmann A., Steinhilber W., Sachs G., Schafer K. Cloning and membrane topology of a P type ATPase from Helicobacter pylori. J. Biol. Chem. 1996;271:446–457. [PubMed: 8550601]
- 67.
- Michetti P., Corthesy-Theulaz I., Davin C., Haas R., Vaney A.-C., Heitz M., Bille J., Kraehenbuhl J.-P., Saraga E., Blum A. L. Immunization of BALB/c mice against Helicobacter felis infection with Helicobacter pylori urease. Gastroenterology. 1994;107:1002–1011. [PubMed: 7926454]
- 68.
- Michetti P., Kreiss C., Kotloff K. L., Porta N., Blanco J. L., Bachmann D., Herranz M., Saldinger P. F., Corthesy-Theulaz I., Losonsky G., Nichols R., Simon J., Stolte M., Ackerman S., Monath T. P., Blum A. L. Oral immunization with urease and Escherichia coli heat-labile enterotoxin is safe and immunogenic in Helicobacter pylori-infected adults. Gastroenterology. 1999;116:804–812. [PubMed: 10092302]
- 69.
- Mobley H. L. T., Cortesia M. J., Rosenthal L. E., Jones B. D. Characterization of urease from Campylobacter pylori. J. Clin. Microbiol. 1988;26:831–836. [PMC free article: PMC266469] [PubMed: 3384908]
- 70.
- Mobley H. L. T., Garner R. E., Bauerfeind P. Helicobacter pylori nickel transport gene nixA: synthesis of catalytically active urease in E. coli independent of growth conditions. Mol. Microbiol. 1995;16:97–109. [PubMed: 7651142]
- 71.
- Mobley H. L. T., Island M. D., Hausinger R. P. Molecular biology of microbial ureases. Microbiol. Rev. 1995;59:451–480. [PMC free article: PMC239369] [PubMed: 7565414]
- 72.
- Moore R. A., Kureishi A., Wong S., Bryan L. E. Categorization of clinical isolates of Helicobacter pylori on the basis of restriction digest analysis of PCR-amplified ureC genes. J. Clin. Microbiol. 1993;31:1334–1335. [PMC free article: PMC262931] [PubMed: 8099088]
- 73.
- Mulrooney S. B., Hausinger R. P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J. Bacteriol. 1990;172:5837–5843. [PMC free article: PMC526901] [PubMed: 2211515]
- 74.
- Myers G. A., Ermak T. H., Georgakopoulos K., Tibbitts T., Ingrassia J., Gray H., Kleanthous H., Lee C. K., Monath T. P. Oral immunization with recombinant Helicobacter pylori urease confers long-lasting immunity against Helicobacter felis infection. Vaccine. 1999;17:1394–1403. [PubMed: 10195775]
- 75.
- Neyrolles O., Ferris S., Behbahani N., Montagnier L., Blanchard A. Organization of Ureaplasma urealyticum urease gene cluster and expression in a suppressor strain of Escherichia coli. J. Bacteriol. 1996;178:647–655. [PMC free article: PMC177707] [PubMed: 8550495]
- 76.
- Nicholson E. B., Concaugh E. A., Foxall P. A., Island M. D., Mobley H. L. T. Proteus mirabilis urease: transcriptional regulation by ureR. J. Bacteriol. 1993;175:465–473. [PMC free article: PMC196161] [PubMed: 7678244]
- 77.
- Nolan, K., D. J. McGee, H. M. Mitchell, T. Kolesnikow, J. M. Harro, J. O'Rourke, J. E. Wilson, N. D. Moss, H. L. T. Mobley, and A. Lee. In vivo behaviour of Helicobacter pylori SS1 nixA mutant with reduced urease activity. Submitted for publication. [PMC free article: PMC127660] [PubMed: 11796600]
- 78.
- Owen R. J., Bickley J., Hurtado A., Fraser A., Pounder R. E. Comparison of PCR-based restriction length polymorphism analysis of urease genes with rRNA gene profiling for monitoring Helicobacter pylori infections in patients on triple therapy. J. Clin. Microbiol. 1994;32:1203–1210. [PMC free article: PMC263645] [PubMed: 7914204]
- 79.
- Pappo J., Thomas W. D. Jr, Kabok Z., Taylor N. S., Murphy J. C., Fox J. G. Effect of oral immunization with recombinant urease on murine Helicobacter felis gastritis. Infect. Immun. 1995;63:1246–1252. [PMC free article: PMC173142] [PubMed: 7890380]
- 80.
- Park I.-S., Hausinger R. P. Site directed mutagenesis of Klebsiella aerogenes urease: identification of histidine residues that appear to function in nickel ligation, substrate binding, and catalysis. Protein Sci. 1993;2:1034–1041. [PMC free article: PMC2142404] [PubMed: 8318888]
- 81.
- Park I.-S., Hausinger R. P. Requirement of CO2 for in vitro assembly of the urease nickel metallocenter. Science. 1995;267:1156–1158. [PubMed: 7855593]
- 82.
- Phadnis S. H., Parlow M. H., Levy M., Ilver D., Caulkins C. M., Connors J. B., Dunn B. E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 1996;64:905–912. [PMC free article: PMC173855] [PubMed: 8641799]
- 83.
- Reitzer, L. J., and B. Magasanik. 1987. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 302–320. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, 1st ed. American Society for Microbiology, Washington, D.C.
- 84.
- Rektorschek M., Buhmann A., Weeks D., Schwan D., Bensch K. W., Eskandari S., Scott D., Sachs G., Melchers K. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol. Microbiol. 2000;36:141–152. [PubMed: 10760171]
- 85.
- Riddles P. W., Whan V., Blakeley R. L., Zerner B. Cloning and sequencing of a jack bean urease-encoding cDNA. Gene. 1991;108:265–267. [PubMed: 1721034]
- 86.
- Rokita E., Makristathis A., Hirschi A. M., Rotter M. L. Purification of surface-associated urease from Helicobacter pylori. J. Chromatogr. B. Biomed. Sci. Appl. 2000;737:203–212. [PubMed: 10681057]
- 87.
- Sato T., Fujino M. A., Kojima Y., Ohtsuka H., Ohtaka M., Kubo K., Nakamura T., Morozumi A., Nakamura M., Hosaka H. Endoscopic urease sensor system for detecting Helicobacter pylori on gastric mucosa. Gastrointest. Endosc. 1999;49:32–38. [PubMed: 9869720]
- 88.
- Scott D. R., Marcus E. A., Weeks D. L., Lee A., Melchers K., Sachs G. Expression of the Helicobacter pylori ureI gene is required acidic pH activation of cytoplasmic urease. Infect. Immun. 2000;68:470–477. [PMC free article: PMC97165] [PubMed: 10639406]
- 89.
- Scott D. R., Weeks D., Hong C., Postius S., Melchers K., Sachs G. The role of internal urease in acid resistance and Helicobacter pylori. Gastroenterology. 1998;114:58–70. [PubMed: 9428219]
- 90.
- Shirai M., Fujinaga R., Akada J. K., Nakazawa T. Activation of Helicobacter pylori ureA promoter by a hybrid Escherichia coli–H. pylori rpoD gene in E. coli. Gene. 1999;239:351–359. [PubMed: 10548737]
- 91.
- Skouloubris S., Thiberge J. M., Labigne A., De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 1998;66:4517–4521. [PMC free article: PMC108549] [PubMed: 9712811]
- 92.
- Skurnik M., Batsford S., Mertz A., Schiltz E., Toivanen P. The putative arthritogenic cationic 19-kilodalton antigen of Yersinia enterocolitica is a urease β-subunit. Infect. Immun. 1993;61:2498–2504. [PMC free article: PMC280875] [PubMed: 8500886]
- 93.
- Solnick J. V., Canfield D. R., Hansen L. M., Torabian S. Z. Immunization with recombinant Helicobacter pylori urease in specific-pathogen-free rhesus monkeys (Macaca mulatta). Infect. Immun. 2000;68:2560–2565. [PMC free article: PMC97459] [PubMed: 10768944]
- 94.
- Sriwanthana B., Island M. D., Mobley H. L. T. Sequence of the Proteus mirabilis urease accessory gene ureG. Gene. 1993;129:103–106. [PubMed: 8335248]
- 95.
- Suerbaum S., Thiberge J.-M., Kansau I., Ferrero R. L., Labigne A. Helicobacter pylori hspA-hspB heat-shock gene cluster: nucleotide sequence, expression, putative function and imunogenicity. Mol. Microbiol. 1994;14:959–974. [PubMed: 7715457]
- 96.
- Szczebara F., Dhaenens L., Armand S., Husson M. O. Regulation of the transcription of genes encoding different virulence factors in Helicobacter pylori by free iron. FEMS Microbiol. Lett. 1999;175:165–170. [PubMed: 10386365]
- 97.
- Takahashi S., Igarashi H., Nakamura K., Masubuchi N., Saltos S., Aoyagi T., Itoh T., Hirata I. Helicobacter pylori urease activity—comparative study between urease positive and urease negative strain. Jpn. J. Clin. Med. 1993;51:3149–3153. [PubMed: 8283623]
- 98.
- Tsuda M., Karita M., Morshed M. G., Okita K., Nakasaki T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 1994;62:3586–3589. [PMC free article: PMC303000] [PubMed: 8039935]
- 99.
- Turbett G. R., Hoj P. B., Horne R., Mee B. J. Purification and characterization of the urease enzymes of Helicobacter species from humans and animals. Infect. Immun. 1992;60:5259–5266. [PMC free article: PMC258305] [PubMed: 1452359]
- 100.
- van Zwet A. A., Thijs J. C., Kooistra-Smid A. M. D., Schirm J., Snijder J. A. M. Sensitivity of culture compared with that of polymerase chain reaction for detection of Helicobacter pylori from antral biopsy specimens. J. Clin. Microbiol. 1993;31:1918–1920. [PMC free article: PMC265660] [PubMed: 8349775]
- 101.
- Weeks D. L., Eskandara S., Scott D. R., Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. [PubMed: 10642549]
- 102.
- Westblom T. U., Phadnis S., Yang P., Czinn S. J. Diagnosis of Helicobacter pylori infection by means of a polymerase chain reaction assay for gastric juice aspirates. Clin. Infect. Dis. 1993;16:367–371. [PubMed: 8452948]
- Review Recent advances in design of new urease inhibitors: A review.[J Adv Res. 2018]Review Recent advances in design of new urease inhibitors: A review.Kafarski P, Talma M. J Adv Res. 2018 Sep; 13:101-112. Epub 2018 Jan 31.
- Mucosal immunization with a urease B DNA vaccine induces innate and cellular immune responses against Helicobacter pylori.[Helicobacter. 2006]Mucosal immunization with a urease B DNA vaccine induces innate and cellular immune responses against Helicobacter pylori.Hatzifoti C, Roussel Y, Harris AG, Wren BW, Morrow JW, Bajaj-Elliott M. Helicobacter. 2006 Apr; 11(2):113-22.
- Review Ureases as a target for the treatment of gastric and urinary infections.[J Clin Pathol. 2010]Review Ureases as a target for the treatment of gastric and urinary infections.Follmer C. J Clin Pathol. 2010 May; 63(5):424-30.
- Review Bacterial factors that mediate colonization of the stomach and virulence of Helicobacter pylori.[FEMS Microbiol Lett. 2007]Review Bacterial factors that mediate colonization of the stomach and virulence of Helicobacter pylori.Clyne M, Dolan B, Reeves EP. FEMS Microbiol Lett. 2007 Mar; 268(2):135-43.
- Urea, fluorofamide, and omeprazole treatments alter helicobacter colonization in the mouse gastric mucosa.[Helicobacter. 2006]Urea, fluorofamide, and omeprazole treatments alter helicobacter colonization in the mouse gastric mucosa.Aristoteli LP, O'Rourke JL, Danon S, Larsson H, Mellgard B, Mitchell H, Lee A. Helicobacter. 2006 Oct; 11(5):460-8.
- Urease - Helicobacter pyloriUrease - Helicobacter pylori
Your browsing activity is empty.
Activity recording is turned off.
See more...