This chapter describes glycosylation in the Arthropoda, focusing primarily on Drosophila melanogaster. The major glycan classes are similar to those described in vertebrates, with interesting differences. The powerful genetic systems available for studying gene function in D. melanogaster have proved to be effective means for understanding glycan function in early development and have provided some of the first examples of how glycans affect growth factor signaling, morphogen gradients, protein secretion, and neural function in vivo.
HISTORICAL PERSPECTIVE
Arthropods are among the most successful species on Earth and are found in all types of environments. One of their characteristic features is an exoskeleton composed of chitin, which provides support and physical protection. The best-studied example is the fruit fly, D. melanogaster. In 1910, T.H. Morgan published the first paper about the genetics of D. melanogaster, which showed that white eye color was a sex-linked trait. Since then, this organism has been the predominant model organism for genetic analysis in animals. Its advantages include an easily studied developmental program, a sequenced and actively annotated genome, a relatively complex neural system, and the ability to discern literally thousands of different phenotypes in morphology, development, and behavior.
In pursuit of genes that regulate development, many Drosophila geneticists ran head-on into glycans. New analytical techniques have expanded the appreciation for the glycan synthetic capacity of the organism and have helped establish links between interesting phenotypes and altered glycan expression. Some of these associations have proven to be common across species and others are unique to D. melanogaster. Given the complexity of glycosylation in Drosophila, it is impossible to cover all aspects of glycans and glycan-binding proteins in this organism. Instead, we provide an overview of the major classes of arthropod glycans and examples of how studying glycans in Drosophila can lead to new discoveries that impact vertebrate as well as invertebrate biology.
INSECT GLYCOPROTEINS
N-Linked Glycan Assembly and Diversity
Although it was once thought that arthropod glycoproteins were exclusively of the high-mannose or paucimannose type (Chapter 9), annotation of the D. melanogaster genome predicted the existence of the enzymatic machinery needed to generate hybrid and complex glycans. Moreover, improved analytic techniques allowed the detection of very minor glycans. The commercial and experimental demand for eukaryotic expression systems led to the characterization of the glycosylation pathways of cells derived from the moth Spodoptera frugiperda (Sf9 cells) and from D. melanogaster (S2 cells). It is now clear that high-mannose and paucimannose glycans account for >90% of the total N-linked glycan diversity in Drosophila and other insects throughout their life cycles. However, hybrid and complex glycans, including sialylated, sulfated, glucuronylated, and zwitterionic structures, are also present, albeit as minor components (Figure 26.1).
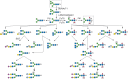
FIGURE 26.1.
N-Linked glycan diversity in Drosophila and other insects. N-Linked glycan processing after endoplasmic reticulum (ER) mannosidase trimming to the Man5GlcNAc2 structure is shown. (Gray boxes) The predominant N-linked glycans, Man5GlcNAc2 and Man3GlcNAc (more...)
Drosophila adds fucose (Fuc) in both α1-3 and α1-6–linkages to the reducing terminal N-acetylglucosamine, whereas vertebrates restrict this linkage to α1-6. Fucα1-3GlcNAc is immunogenic in humans and rabbits, resulting in the production of antibodies against the so-called horseradish peroxidase (HRP) epitope. Anti-HRP antibodies show that the Fucα1-3GlcNAc epitope is restricted primarily to neural tissue in a broad range of arthropods. Drosophila does not extend its core fucose residues with additional capping monosaccharides nor modify its N-linked glycans by O-methylation, both of which occur extensively in Caenorhabditis elegans (Chapter 25). The demonstration of fucosylated, sialylated, sulfated, hybrid, biantennary complex, and triantennary complex glycans in all stages of the D. melanogaster life cycle makes the diversity of the arthropod N-glycans generally comparable to that of mammals (Chapter 27), except for the very limited use of sialic acids (Sias).
The relative paucity of complex N-linked glycans in Drosophila has been attributed at least partially to the presence of a hexosaminidase in the secretory pathway. Encoded by the fused lobes (fdl) gene, the enzyme is capable of efficiently removing N-acetylglucosamine residues that are added by N-acetylglucosaminyltransferase I (GlcNAcT-I encoded by Mgat1) to the nonreducing terminal Manα1-3 arm of the Man5GlcNAc2 core glycan. GlcNAcT-I catalyzes the first committed step toward the generation of hybrid or complex glycans (Chapter 9). Therefore, removal of this N-acetylglucosamine residue effectively blocks subsequent extension reactions, generating the observed predominance of high-mannose or paucimannose glycans on glycoproteins.
The presence of hybrid and complex glycans in Drosophila predicts the existence of specific enzymes that act on acceptor substrates that have escaped trimming by Fdl. For instance, α1-6 fucosyltransferase, GlcNAcT-I, -II, -III, -IV, galactosyltransferase (GalT), and sialyltransferase (SiaT) activities are required to generate structures more complex than paucimannose glycans. Of these enzymes, only GlcNAcT-I and the single SiaT are reasonably well-characterized in Drosophila. Genome annotations predict the existence of the other enzymes, although their activity and expression have not been characterized.
Phenotypes have been described for mutations in key N-linked glycan processing and synthetic enzymes (Table 26.1). Loss of the Golgi-trimming enzyme, α1-2-mannosidase (Golgi mannosidase I, mas1) has little or no impact on the processing of high-mannose glycans, which led to the identification of an alternative mannosidase activity that effectively bypasses loss of mannosidase I. However, the embryonic peripheral nervous system, the wing, and the adult eye show mild alterations in the Drosophila mas1 mutant, suggesting that the bypass is not complete in all tissues, similar to the tissue-specific bypass identified in mouse following targeted disruption of mouse α-mannosidase (mGMII, Golgi mannosidase II) (Chapter 9). To date, relatively few mutations in genes affecting core N-linked glycosylation have been reported in Drosophila. One exception is the glucosyltransferase Alg5, which adds glucose (Glc) to the Dol-P-GlcNAc2Man9 precursor. Mutations in the Drosophila Alg5 gene, known as wollknäuel (wol), show deficient protein secretion, extracellular matrix deposition, and cuticle elaboration, phenotypes that reflect inefficient protein glycosylation and altered endoplasmic reticulum (ER) function.
TABLE 26.1.
Genes that affect the synthesis or function of Drosophila glycans
Mutations in genes responsible for subsequent processing steps have begun to reveal the importance of glycan complexity in Drosophila. Reduction in the activity of the Fdl hexosaminidase results in altered brain structure. Brain lobes, normally separated in wild-type adults, are fused together through a continuous stalk at the midline in fdl mutants, hence, the original name of the mutation, fused lobes (Figure 26.2). In the wild-type adult, the separated lobes form portions of the mushroom body, a brain structure whose function has been implicated in Drosophila learning and memory. In an interesting convergence, a null mutation in Mgat1 generates an apparently identical fused-lobes phenotype, although the glycan expression profile shifts from more complex in fdl mutants to high mannose and paucimannose in Mgat1 mutants. It is still unclear how loss of complex glycans in one case (Mgat1) or enrichment of complex glycans in another case (fdl) generates the same neural phenotype. Mgat1 mutants also show reduced locomotor activity and decreased life span. The decreased life span is rescued by re-expression of the enzyme in neural tissue alone, indicating an essential role for complex glycosylation in neural functions that impact whole organism physiology.
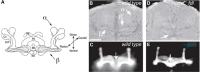
FIGURE 26.2.
Mutations in enzymes that process complex N-linked glycans alter adult brain morphology in D. melanogaster. (A) The major lobes of the adult brain are shown in cross section. (Hatched areas) γ-lobes; (α,β) α- and β-lobes; (more...)
A terminal synthetic step in the production of N-linked glycans in mammals is the addition of sialic acid residues. Genomic annotation and biochemical studies have shown that Drosophila possesses some, but not all, of the biosynthetic machinery necessary for the production of CMP-NeuAc. A single SiaT identified in Drosophila shows a two-fold preference for transferring sialic acid to a LacdiNAc (GalNAcβ1-4GlcNAc) acceptor compared to a type II LacNAc (Galβ1-4GlcNAc) acceptor. However, the only identified sialylated N-linked glycans in Drosophila possess subterminal LacNAc instead of LacdiNAc, despite the presence of LacdiNAc terminated N-linked glycans in the organism. SiaT is detected in a very small number of neurons beginning in late embryonic stages, but expands to a larger number of central nervous system (CNS) neurons in larval and adult stages. The relatively restricted expression of the SiaT enzyme reinforces proposals that minor glycans may be restricted to small subsets of cells in specific developmental or adult stages (Chapter 7). In fact, loss of glycoprotein sialylation results in explicit neurologic defects in adult flies, including behavioral abnormalities and temperature-induced seizures, indicating the importance of this minor modification of complex glycans for normal neurophysiological function.
O-Linked Glycan Assembly and Diversity
Insects add glycans in O-linkage to serine and threonine residues on secreted, cell-surface, and intracellular proteins. Structural complexity ranges from single monosaccharides (N-acetylgalactosamine, N-acetylglucosamine, mannose, glucose, or fucose) to extensively modified glycosaminoglycan chains) (see below). The core-1 structure (Galβ1-3GalNAcα-Thr/Ser), which is found extensively on vertebrate mucin-like proteins (Chapter 10), has also been described in insects (Figure 26.3). As much as 40% of the total mass of these proteins is contributed by glycans. In Drosophila tissues, peanut agglutinin lectin and mucin-specific antibodies reveal developmentally regulated and spatially restricted expression of Galβ1-3GalNAcα-O-protein moieties.
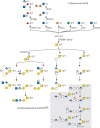
FIGURE 26.3.
O-Linked glycan diversity in Drosophila and other insects. Genetic and biochemical analysis of Drosophila mutants were instrumental in defining developmental roles for domain-specific, simple mono-, di- or trisaccharide modifications on serine/threonine (more...)
Drosophila possesses a family of 10 polypeptide N-acetylglucosaminyltransferases (ppGalNAcT/PGANTs encoded by pgant genes; Chapter 10). Systematic mutational and RNAi-mediated knockdown analysis of individual pgant genes showed that pgant4, 5, 7, and 35A are essential for viability. Loss of pgant3 impacts extracellular matrix secretion and integrin-mediated cell adhesion during development, and pgant4 modulates proteolytic cleavage of substrates essential for secretory granule formation. Loss of pgant4 also results in loss of the secreted peritrophic membrane of the digestive tract, leading to disruption of the progenitor cell niche. pgant9 affects glycosylation of certain mucins and the morphology of secretory granules. Thus, mucin-type O-glycosylation in Drosophila regulates essential developmental programs and modulates trafficking through the secretory pathway, providing blueprints for studying similar functions in vertebrate systems.
Once initiated by ppGalNAcT/PGANT enzymes, the extension of mucin type glycans in Drosophila results in the production of three different core structures. Core 1 structures predominate, but core 2 glycans are also present, as well as an incompletely defined HexNAc-HexNAc core. A family of core 1 galactosyltransferases (C1GalTs) exists in Drosophila and the loss of certain members disrupts the nervous system and neuromuscular junctions (NMJ). Each core type can be modified by the addition of glucuronic acid (GlcA), either as a terminal or as a branching residue. Loss of the glucuronic acid transferase GlcAT-P results in the loss of glucuronic acid on core 1 structures and results in nervous system defects and alterations in the NMJs. Glucuronic acid is also detected as an internal residue within an extended chain, reminiscent of a glycosaminoglycan disaccharide structural unit (Chapter 17) or of the matriglycan repeat on mammalian dystroglycan (Chapter 13). In Drosophila and mosquito, sulfated mucin type glycans are also detected. Species-specific expression also exists with regard to the relative abundance of different core structures and the existence of specific postsynthetic modifications. For example, O-glycans can be decorated with phosphoethanolamine linked to HexNAc residues in mosquito and wasp species, but not in Drosophila.
Other types of O-linked glycan modifications are also present in insects. The discovery that O-GlcNAc decorates protein components of the Drosophila polytene chromosome added weight to initial demonstrations of the existence of nucleocytoplasmic glycosylation in animal cells (Chapters 18 and 19). Subsequently, this finding was reinforced by the observation that the gene encoding the O-GlcNAc transferase (OGT), was affected in super sex combs (sxc) mutants. The sxc locus is one of several polycomb group (PcG) genes that function as homeotic regulators of gene expression along the antero-posterior axis during Drosophila development. In a dramatic convergence of independent studies, major sites of PcG binding along the Drosophila genome were also shown to be major sites of genomic O-GlcNAc modification, indicating that addition of O-GlcNAc to DNA-binding proteins regulates their genomic binding specificity. Additionally, O-GlcNAc is also added to secreted and membrane proteins by the extracellular O-GlcNAc transferase EOGT. O-GlcNAc has been found on the extracellular matrix protein Dumpy, as well as Notch and Notch ligands. Loss of Eogt results in larval lethality.
O-linked mannose, a modification that contributes to the pathophysiology of some human muscular dystrophies (Chapters 13 and 45), is also present in Drosophila and is added to proteins by two transferase genes. Mutations in either of these two genes, rotated abdomen (rt) or twisted (tw), cause clockwise helical rotations of abdominal morphology in adult flies. The two genes interact such that the abdominal rotation observed in compound hypomorphic mutants is more severe than that of either single mutant. However, their alleles also show mutually epistatic interactions and their strong loss-of-function alleles are phenotypically indistinguishable, suggesting that Rt and Tw proteins function in a complex. Rt and Tw proteins must be expressed together in the same cell to achieve transfer of mannose onto dystroglycan, a physiologic acceptor for O-mannosylation.
Addition of a simple glycan, O-linked fucose, modulates complex developmental signals. The Notch gene was first identified and characterized in Drosophila as encoding a large, multimodular receptor protein involved in cell-fate determination. One characteristic of Notch is the presence of epidermal growth factor (EGF)-like modules. The EGF-like domain modules in the human Notch homolog have been shown to contain three domain-specific types of O-linked glycosylation (O-Glc, O-Fuc, and O-GlcNAc), which have been implicated in various steps in Notch signaling, including Notch folding, cleavage and interactions with its ligands, Delta and Serrate/Jagged (Chapter 13). As in mouse and humans, Drosophila possesses a protein O-glucosyltransferase, named Rumi, and two protein O-fucosyltransferases, O-fut1 and O-fut2. As in vertebrates, Drosophila O-fut1 transfers fucose to EGF-like domains and O-fut2 transfers fucose to thrombospondin-type repeats on different sets of proteins. Loss of O-fut1 in Drosophila yields Notch mutant phenotypes, indicating that fucosylation of Notch is essential for ligand-induced activation (Figure 26.4A–C). Loss of Rumi also produces Notch-like phenotypes, but in a temperature-sensitive manner, leading to the hypothesis that O-glucosylation is important for cleavage of Notch by appropriate processing enzymes. In Drosophila and vertebrate organisms, O-linked glucose residues are extended by the addition of xylose (Xyl) residues by the xylosyltransferases Shams and Xxylt (Figure 26.3). In addition, Rumi possesses dual nucleotide sugar donor specificity, allowing it to transfer either glucose or xylose to serine (but not threonine) residues in specific sequence contexts.
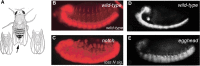
FIGURE 26.4.
Cell fate choices dependent on Notch require appropriate glycan expression. (A) Notch mutations were originally identified based on aberrant wing morphology. Changes in cell fate generate wings that are notched at their margins (arrow). Wing notches arise (more...)
Fucosylated Notch is a substrate for an N-acetylglucosaminyltransferase encoded by the fringe gene. Elongation of O-linked fucose with N-acetylglucosamine yields Notch protein that is more efficiently activated by Delta than by Serrate/Jagged (Chapter 13). Therefore, O-linked glycosylation functions as a switch that activates the Notch receptor and alters its ligand preference. Differential Notch activation by cell-specific expression of O-fut1 and Fringe generates distinct cell fate choices in embryos, larvae, and pupae. The ability of the Drosophila O-fut1 and Fringe proteins to rescue or modify Notch protein or Notch mutant phenotypes is well-documented in cultured cells and in whole embryos, although the demonstration of O-Fuc or the GlcNAc-extended disaccharide on Notch protein extracted from Drosophila tissue has not yet been achieved. In mice and humans, but apparently not in Drosophila, the disaccharide can be extended by the addition of galactose (Gal) and then capped with sialic acid (Chapter 13). Drosophila embryos, larvae, and imaginal tissues also contain GlcNAcβ-3Fuc core structures branched by addition of a glucuronic acid to fucose (Figure 26.3). The functional importance of the glucuronyl branching in Drosophila is not yet known.
INSECT PROTEOGLYCANS AND GLYCOSAMINOGLYCANS
Glycosaminoglycan (GAG) Structure
Drosophila has the complete set of heparan sulfate (HS) biosynthetic and modifying enzymes found in mammalian species, and produces complex HS structures but has only one gene for each class of these enzymes. Disaccharide profiling of GAGs from Drosophila showed the presence of HS and chondroitin sulfate (CS) remarkably similar in structure to that of GAGs found in vertebrates (Chapter 17). The principal disaccharide species of HS-derived units include N-, 2-O, and 6-O sulfated forms, and mono-, di-, and trisulfated disaccharides. CS detected from whole embryos or larvae is largely unsulfated or 4-O-sulfated, but 6-O-sulfated disaccharides have been detected as well. The covalent attachment of GAG chains to proteins via the canonical tetrasaccharide linker (GlcA-Gal-Gal-Xyl) has also been shown for Drosophila. CS and HS have been documented in many other arthropod species, establishing the conservation of these macromolecules throughout this phylum. In contrast, Drosophila and presumably other arthropods do not produce hyaluronan (Chapter 16). Many of the HS-bearing proteoglycan core proteins known from vertebrate systems are also represented in Drosophila. Drosophila has a single Syndecan (Sdc) gene, two glypicans, division abnormally delayed (dally) and dally-like protein (dlp), testican (Cow), and perlecan, which is encoded by the terribly reduced optic lobe (trol) gene. Compared to HSPGs, CSPG core proteins are less well-conserved between species. Therefore, identifying new CSPGs cannot rely on the sequence homology to previously identified vertebrate CSPGs. Recently, glycoproteomic approaches have been successfully used to identify new CSPGs in Drosophila and C. elegans.
Mutations affecting GAG biosynthesis and modification have provided important insights concerning proteoglycan biosynthesis and development. For example, loss of the single N-deacetylase–N-sulfotransferase gene in Drosophila (sfl) results in essentially an unsulfated polymer, N-acetylheparosan, which causes defective patterning decisions orchestrated by several growth factors. Both Ext1- and Ext2-related genes, ttv and sotv, are required for HS polymerization in Drosophila. Mutations affecting the single Hs2st or Hs6st genes have particularly interesting effects. Loss of Hs2st eliminates 2-O sulfate groups as expected, but results in compensatory increases in 6-O-sulfation. The converse is also true: loss of Hs6st produces a polymer lacking 6-O-sulfate groups but 2-O-sulfation and N-sulfation increase, retaining the overall sulfation state of the polymer. Compensation between 2-O and 6-O modifications has also been observed in CHO cell mutants and mouse embryonic fibroblasts derived from Hs2st mutants. HS and CS biosynthesis are also linked in a compensatory fashion; mutations that reduce, but do not eliminate, Ext activity required for HS polymerization increase the net amount of chondroitin polymer formed, similar to observations made in vertebrate systems. These data suggest an important conserved mechanism for retaining the activity of proteoglycans in vivo.
Morphogen Signaling and Organ Size Control
Genetic studies in Drosophila showed that GAGs are critical for signaling mediated by a number of growth factors during development. HS proteoglycans serve as coreceptors for many growth factors, including Wingless (Wg; a Wnt ortholog), Decapentaplegic (Dpp; a BMP4 ortholog), fibroblast growth factor (FGF), Hedgehog (Hh), and Unpaired (Upd; a ligand of the Drosophila Jak/Stat pathway), and affect their distribution and signaling (Table 26.1). For example, patches of cells that express Dally at high levels showed markedly elevated levels of Dpp signaling in a cell-autonomous manner. Overexpression of Dally may increase Dpp protein levels on the cell surface by disrupting receptor-mediated internalization and degradation.
The ability of HS proteoglycans to alter the distribution and levels of growth factors in the matrix has biological significance because many of these secreted proteins are morphogens, secreted protein factors that show graded distributions across tissues and provide an essential mechanism for generating cell diversity during tissue assembly. Morphogen activity has been shown in vivo for Wg, Dpp, and Hh and Upd in Drosophila. HS proteoglycans not only affect their levels in tissues, but are also integrated into regulatory circuits of the morphogen systems. For example, both expression of a Dpp receptor (Thickveins) and Dally, a coreceptor for Dpp, are negatively regulated by Dpp signaling. This feedback system provides a mechanism to adjust cellular responses in the face of inappropriate reductions in Dpp signaling that might arise from genetic or environmental perturbations.
Recent studies have highlighted the role of the glypicans in organ size control in the context of evolutionary developmental biology. Halteres, which contribute to body balance during flight, are characteristic of two-winged Dipteran species, like Drosophila, and have evolved from the hindwings of four-winged ancestral species. This knob-like structure is much smaller than the ancestral wing from which it evolved. The size difference between the two appendages is controlled by a Hox gene, Ultrabithorax (Ubx), which regulates haltere identity. In the haltere disc, Ubx directly represses expression of the dally gene, limiting the bone morphogenetic protein (BMP) signaling and, thereby, organ size. Hox control of morphogen signaling through HS proteoglycan gene expression is one of the general mechanisms used across animal evolution to modify homologous organ structures.
Proteoglycans in the Stem Cell Niche
HS proteoglycans are an evolutionarily conserved, universal component of stem cell niches. In the ovarian germline stem cell (GSC) niche, Dally regulates stem cell number by acting as a niche size determinant. The cells that constitute the niche produce Dpp, which acts on a GSC that is in direct contact with a niche cell. One of the daughter cells arising from the division of a GSC will retain this contact and maintain its stem cell character. The other daughter cell, which loses this contact, will differentiate. Dally expressed in niche cells acts as a “trans” coreceptor, stabilizing Dpp and presenting Dpp to GSCs that are in direct contact with the niche cell (Figure 26.5). In dally mutants, Dpp signaling in GSCs is impaired and GSCs are lost to differentiation. In contrast, dally overexpression in a somatic cell population outside the niche results in ectopic activation of Dpp signaling in germ cells in this region, leading to an expansion of GSC-like cells. Thus, contact-dependent signaling by HS proteoglycans provides a mechanism to define the physical space of the niche and to control stem cell number.
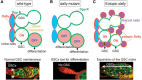
FIGURE 26.5.
Glycosaminoglycans regulate the contact-dependent maintenance of germline stem cells (GSCs). (A) The niche cells (blue) express Dally on the cell surface (red) and germ cells express Dpp receptors (green). A dividing GSC is shown at the top and two daughter (more...)
Proteoglycans in Neural Development
The essential requirement for proteoglycans in neural development has also been illuminated by genetic analysis in Drosophila. Syndecan (Sdc) regulates axon guidance by modulating Slit-Robo signaling in the embryonic CNS. Dally-like (Dlp) also influences nervous system development but has distinct functions from Sdc. Sdc affects axon guidance decisions near the midline of the nervous system, whereas Dlp is required for fascicle formation at a distance from the midline. Trol regulates embryonic motor axon guidance by facilitating transmembrane signaling mediated by Semaphorin–Plexin interactions. HS proteoglycans also regulate synaptogenesis at the neuromuscular junction (NMJ). Both Sdc and Dlp bind to LAR, a receptor tyrosine phosphatase, which controls NMJ growth and active zone morphogenesis. As in axon guidance, these two proteoglycans have distinct functions: Sdc promotes the growth of presynaptic terminals, whereas Dlp regulates active zone formation. Trol regulates the formation of both pre- and postsynaptic structures by localizing Wg protein.
CHITIN
Chitin, a polymer of (GlcNAcβ4)n, is one of the most abundant biopolymers on Earth, second only to cellulose (which is interestingly a (Glcβ4)n polymer synthesized in a similar manner by extrusion at the cell surface). Chitin is a major component of the rigid, cuticular exoskeleton of all Arthropoda, and therefore these animals expend considerable resources toward its assembly. Chitin fibrils also form a more subtle, protective layer at the apical surface of gut epithelial cells (the peritrophic membrane) and in the lumen of the forming tracheal system. Elegant genetic screens have shown that mutations in genes necessary for chitin polymerization, modification, and disassembly affect tracheal morphology (Table 26.1). The potential for chitin or for small chitin-based oligosaccharides to modulate cell signaling and morphogenesis remains unexplored in Drosophila.
INSECT GLYCOSPHINGOLIPIDS
Insects were once classified as “animals without gangliosides” in reference to sialylated glycosphingolipids (GSLs) found broadly distributed in animal families other than the arthropods (Chapter 11). However, arthropods possess their own family of GSLs, designated as the arthroseries. Sialylated GSLs (gangliosides) have not been found in Drosophila, but the arthroseries glycolipids contain glucuronic acid rather than sialic acid, and like many of the ganglioside sialic acids, the glucuronic acid is linked to a terminal galactose residue (Figure 26.6).
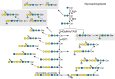
FIGURE 26.6.
Glycosphingolipid glycan diversity. The arthroseries glycosphingolipids (GSLs) are built by extension from a mactosylceramide core (Manβ1-4Glcβ-ceramide). Neutral arthroseries glycans are frequently modified by the addition of phosphoethanolamine (more...)
Vertebrates generally build GSLs on a lactosylceramide core (Galβ1-4Glcβ-ceramide). In contrast, the arthropods add a mannose residue to Glcβ-Cer to generate a core (Manβ1-4Glcβ-ceramide), called “mactosylceramide.” Addition of the next two monosaccharides, N-acetylglucosamine in β1-3 linkage to the underlying mannose and a terminal N-acetylgalactosamine in β1-4 linkage, produces a substrate for the addition of ethanolamine-phosphate (EthNP) to C-6 of N-acetylglucosamine (EthNP-6GlcNAcβ1-3Manβ1-4Glcβ-Cer). Therefore, the neutral core of most arthroseries glycolipids is more correctly described as zwitterionic rather than neutral. Arthroseries cores with more than one N-acetylglucosamine residue are found with zero, one, or two EthNP groups. The functional significance of EthNP modification is currently unknown.
Additional species-specific core diversity has also been identified. For example, the arthroseries triosylceramide in Drosophila can be alternatively extended with galactose rather than N-acetylgalactosamine, followed by addition of glucuronic acid (Figure 26.6). Other arthropod species add galactose to mactosylceramide or to the arthroseries tetraosylceramide with subsequent capping and EthNP modification (Figure 26.6). The expanding diversity of alternative core extensions detected in arthropods provides a larger pool of structural variants from which these organisms can tailor GSL expression for specific developmental or tissue-restricted functions, similar to core switching in vertebrate species (Chapter 11).
Mutations that affect the first steps in Drosophila GSL synthesis were originally identified as modulators of cell fate. Biochemical analysis showed that genes called egghead (egh) and brainiac (brn), respectively, encode the mannosyltransferase and N-acetylglucosaminyltransferase that add the second and third monosaccharide residues (Figure 26.6). Phenotypes associated with loss of maternal and zygotic egh and brn include overproliferation of neural cells at the expense of epithelial lineages. These egh/brn phenotypes closely resemble the embryonic phenotype of mutations in the Notch gene (Figure 26.4). An additional phenotype of egh mutants is the formation of enlarged peripheral nerves due to glial cell overproliferation with immune cell infiltration, reminiscent of the human disorder neurofibromatosis type 1. As in this human disorder, overgrowth results from enhanced phosphatidylinositol-3-kinase activity, likely because of reduced Ras signaling. The neurofibromatosis-like phenotypes indicate the importance of the membrane glycolipid environment for propagating appropriate transmembrane signals. Mutations affecting the two partially redundant N-acetylgalactosaminyltransferases that generate the tetraosylceramide of the arthroseries result in subtle behavioral phenotypes in larvae and altered EGF receptor signaling in ovarian follicle cells, but otherwise produce viable, fertile adults. It remains to be determined whether arthroseries glycans elongated beyond the tetraosylceramide possess structural information that imparts specific function.
INSECT LECTINS
Annotation of the Drosophila genome indicates the presence of representatives for each of the known classes of animal lectins (Chapter 28). Intracellular carbohydrate-binding proteins, which in vertebrates are associated with protein folding and quality control in the ER or with trafficking through the early compartments of the secretory pathway (calnexin, calreticulin, VIP, ERGIC-53), are also found in Drosophila (Chapter 39). Further downstream, lysosomal targeting is mediated by LERP, a sorting protein functionally similar to the vertebrate mannose-6-phosphate receptor (Chapter 33). Although LERP possesses domain architecture and sequence similarity to the P-type lectin sequences of the mannose-6-phosphate receptor, its binding does not appear to be glycan-dependent, suggesting evolutionary divergence in the mechanisms that control the biogenesis of lysosomes. The binding specificities and functions of insect family members of the Galectin (Chapter 36), C-type (Chapter 34), and I-type lectin families (Chapter 35) are not well-characterized. For example, the protein encoded by the furrowed gene, a putative C-type lectin with significant homology with vertebrate selectins, has been implicated in planar cell polarity dynamics during eye development, but this activity is independent of its carbohydrate recognition domain (CRD).
Current classification schemes for animal lectins reflect the long history of studying glycan-binding proteins in vertebrates (Chapter 28). However, a handful of lectin activities, first identified in Drosophila, challenge the comprehensiveness of current animal lectin designations. One of these was identified in a search for secreted proteins that stimulate the proliferation and motility of imaginal disc cells. The imaginal disc growth factor (IDGF) family members are structurally related to chitinases but lack amino acid residues essential for catalytic activity. It has been suggested that the IDGF family has evolved away from hydrolysis while maintaining a glycan-binding activity that facilitates mitogenic and trophic support. Another currently unclassified lectin, called “gliolectin,” was identified in a screen of Drosophila cDNAs for proteins that mediate cell adhesion to immobilized glycans. Gliolectin is expressed in a subset of embryonic glial cells found at the midline of the developing CNS and in cells at the dorsal/ventral boundary of the wing imaginal disc. Mutants that lack gliolectin show defects in axonal pathfinding, consistent with a role for glycan-mediated cell adhesion in facilitating the transmission of signals between cells. They also show Notch-like phenotypes in wing development, indicating a broader role for gliolectin in establishing tissue boundaries. Finally, the mind-the-gap gene encodes a protein (MTG) that possesses domain folds consistent with its ability to bind N-acetylglucosamine, although a definitive structural or functional homolog among well-defined lectin families is yet to be determined. In Drosophila larvae, MTG participates in organizing the glycoprotein matrix at the larval neuromuscular junction.
INSECT NUCLEOTIDE SUGAR TRANSPORTERS
Genetic and biochemical approaches have identified multiple nucleotide sugar transport activities in Drosophila. For several of these genes, glycomic, and other phenotypic consequences associated with knockdown or loss-of-function have been characterized (Table 26.1). For others, further analysis is needed to define their specificity and function. The first nucleotide sugar transporters identified in Drosophila were recovered as mutants from genetic screens that targeted phenotypes associated with altered morphogen or growth factor signaling and, therefore, impacted GAG expression. Fringe connection (frc) and Slalom (sll) are responsible for the transport of UDP-GlcA, UDP-GlcNAc, UDP-Xyl, and the sulfation donor PAPS, each of which is essential for GAG biosynthesis. Other transport activities that function in important developmental pathways include GFR/Nac and Efr, which transport GDP-fucose into the Golgi and ER, respectively. Loss of these transport activities impacts Notch signaling and neural-specific glycan expression. Other transporter genes are also associated with developmental phenotypes but their transport specificities require further analysis (Table 26.1).
ACKNOWLEDGMENTS
The authors acknowledge helpful contributions from Hamed Jafar-Najad and Iain B.H. Wilson.
FURTHER READING
- Wiegandt H. 1992. Insect glycolipids. Biochim Biophys Acta 1123: 117–126. doi:10.1016/0005-2760(92)90101-z [PubMed: 1739742] [CrossRef]
- Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. 2007. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem 282: 9127–9142. doi:10.1074/jbc.M606711200 [PubMed: 17264077] [CrossRef]
- Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. 2008. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem 283: 30385–30400. doi:10.1074/jbc.M804925200 [PMC free article: PMC2573061] [PubMed: 18725413] [CrossRef]
- Ten Hagen KG, Zhang L, Tian E, Zhang Y. 2009. Glycobiology on the fly: developmental and mechanistic insights from Drosophila. Glycobiology 19: 102–111. doi:10.1093/glycob/cwn096 [PMC free article: PMC2722416] [PubMed: 18824561] [CrossRef]
- Yan D, Lin X. 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol 1: a002493. doi:10.1101/cshperspect.a002493 [PMC free article: PMC2773635] [PubMed: 20066107] [CrossRef]
- Crickmore M, Mann RS. 2010. A new chisel for sculpting Darwin's endless forms. Nat Cell Biol 12: 528–529. doi:10.1038/ncb0610-528 [PubMed: 20517301] [CrossRef]
- Tran DT, Zhang L, Zhang Y, Tian E, Earl LA, Ten Hagen KG. 2012. Multiple members of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J Biol Chem 287: 5243–5252. doi:10.1074/jbc.M111.306159 [PMC free article: PMC3285305] [PubMed: 22157008] [CrossRef]
- Nakato H. 2015. Heparan sulfate proteoglycans in the Drosophila ovarian germline stem cell niche. In Glycoscience: biology and medicine (ed. Taniguchi M, et al., editors. ), pp. 825–832. Springer, Tokyo.
- Zhang L, Ten Hagen KG. 2019. O-Linked glycosylation in Drosophila melanogaster. Curr Opin Struct Biol 56: 139−145. doi:10.1016/j.sbi.2019.01.014 [PMC free article: PMC6656608] [PubMed: 30852302] [CrossRef]
- Nishihara S. 2020. Functional analysis of glycosylation using Drosophila melanogaster. Glycoconj J 37: 1−14. doi:10.1007/s10719-019-09892-0 [PubMed: 31773367] [CrossRef]
Publication Details
Author Information and Affiliations
Authors
Kelly G. Ten Hagen, Hiroshi Nakato, Michael Tiemeyer, and Jeffrey D. Esko.Copyright
The content of this book is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 Unported license. To view the terms and conditions of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
Publisher
Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY)
NLM Citation
Ten Hagen KG, Nakato H, Tiemeyer M, et al. Arthropoda. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology [Internet]. 4th edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. Chapter 26. doi: 10.1101/glycobiology.4e.26





