This work is distributed under the terms of a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 license (CC BY-NC-ND), a copy of which is available at https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
In this report, we address a scenario that uses synthetic genotype case-control data that is influenced by environmental factors in a genome-wide association study (GWAS) context. The precise way the environmental influence contributes to a given phenotype is typically unknown. Therefore, our study evaluates how to approach a GWAS that may have an environmental component. Specifically, we assess different statistical models in the context of a GWAS to make association predictions when the form of the environmental influence is questionable. We used a simulation approach to generate synthetic data corresponding to a variety of possible environmental-genetic models, including a “main effects only” model as well as a “main effects with interactions” model. Our method takes into account the strength of the association between phenotype and both genotype and environmental factors, but we focus on low-risk genetic and environmental risks that necessitate using large sample sizes (N = 10,000 and 200,000) to predict associations with high levels of confidence. We also simulated different Mendelian gene models, and we analyzed how the collection of factors influences statistical power in the context of a GWAS. Using simulated data provides a “truth set” of known outcomes such that the association-affecting factors can be unambiguously determined. We also test different statistical methods to determine their performance properties. Our results suggest that the chances of predicting an association in a GWAS is reduced if an environmental effect is present and the statistical model does not adjust for that effect. This is especially true if the environmental effect and genetic marker do not have an interaction effect. The functional form of the statistical model also matters. The more accurately the form of the environmental influence is portrayed by the statistical model, the more accurate the prediction will be. Finally, even with very large samples sizes, association predictions involving recessive markers with low risk can be poor.
Acknowledgments
This research was supported by the RTI Fellows Program. The authors acknowledge Craig Hollingsworth and Joanne Studders for their help in improving the readability of the report.
Introduction
In recent years, scientists and researchers have increasingly used the genome-wide association study (GWAS) in attempts to unravel the genetic factors that influence important phenotypes such as disease presence and predisposition. The hypothesis GWAS implies is that if genetic variations are more frequent in people with a given disease, the variations are likely associated with the disease. In general, GWAS apply univariate statistical tests to each gene marker or single nucleotide polymorphism (SNP) as an initial step. This SNP-based test is statistically straightforward, and the core tests for assessing the associations are standard methods (e.g., Chi Square tests, regression) that have been studied outside of and within the GWAS context. Kuo and Feingold1 describe the most commonly used statistical methods applied to GWAS. All the tests they cite are single-locus tests. However, in an earlier paper2 we recommended combining two or more statistical tests if the genetic inheritance properties are not known.
The popularity of the GWAS approach is testimony to its simplicity; however, it obscures the important issue of whether a single-gene model is conducive to unraveling the workings of the biosynthetic pathways of a phenotype. In the path leading from gene to trait, factors such as epigenetics, alternate splicing, gene expression levels, and protein-folding processes create a great deal of complexity.
A number of researchers believe that most complex diseases involve multiple genes and their interactions.3, 4 Although GWAS have had some success in identifying genetic variants underlying complex diseases, most existing studies are based on limited single-locus approaches that detect SNPs based on their marginal associations using a qualitative disease (case-control) diagnosis measure. A further problem with GWAS has been that the genetic (SNP) variation explains only a small proportion of the heritability.5 This issue has been identified in studies of twins, where an alternative estimate of heritability is available.
Researchers can use classical statistical tests derived from case-control experiments to determine whether two loci associate in a GWAS context. Both Pearson’s chi squared test and tests involving logistic regression can be used to examine for pair-wise interaction assumptions. An early study6 investigating gene-gene interactions showed that explicitly modeling interactions between loci for GWAS with hundreds of thousands of markers is computationally feasible. This study also showed that simple methods that explicitly consider interactions can actually achieve reasonably high power with realistic sample sizes under different interaction models with some marginal effects. This is true even after adjusting for multiple testing using the Bonferroni correction. However, the genotype-phenotype scenarios addressed by this study had atypically large effects.
In our study, we focused on low-effect loci with low relative risks of association with disease diagnosis, because the evidence7 suggests these are common. Most GWAS report only small changes in disease risk (1.1 to 1.5). It has also been reported8 that relative risks underestimate the true risk and the corresponding effect size.
The word risk can have a variety of meanings. In an environmental context, it means “a hazard based on an exposure” to a chemical or pollutant such as tobacco smoke. In another context, risk is interpreted more narrowly to mean the probability of an adverse consequence, for example, an adverse event such as a disease. The term environmental risk in this study is used broadly; we define it as any process that contributes to a disease diagnosis that is not genetic in origin. Environmental risks can represent exposure to chemicals or pollutants—or a subject’s age, for example.
Our overarching goal was to identify which statistical methods best identify genotype-phenotype associations when environmental effects also influence the association. Detecting such associations is particularly difficult for genetic variants with modest impacts on risk. Consequently, our experiments specifically investigated scenarios involving low-risk genetic variants and assessed whether environmental influences with varied levels of risk could be a source of the “missing heritability” observed using single-gene models.9 Not surprisingly, our investigations demonstrated that the best statistical method (with respect to statistical power) depends on whether there are interactions between the genotype and environmental factors as well as how well the specified statistical model matches the environmental effect associated with the phenotype. In summary, the simulated dataset provides a truth set for assessing the sensitivity of the effect of the statistical method and of the predicted association. Establishing the genotype-to-phenotype connections without using a simulation approach is difficult to impossible. While our study results demonstrate a number of obvious “truths,” a number of unexpected results may lead researchers to more powerful statistical approaches that can establish the validity of the simulation approach.
Background
Many complex diseases (e.g., diabetes, asthma, cancer) are affected in part by interactions between genes and environmental factors. However, investigators conducting GWAS typically test don’t investigate environmental factors.
There have been several notable exceptions. For example, Terry et al.10 showed a significant interaction between smoking status and the specific gene for lung cancer. Another study, by Stern et al.,11 found smoking status to be an effect modifier of the association between a codon and the risk of bladder cancer. Understanding the relationship between genetic polymorphisms and environmental exposures can greatly aid investigators in detecting high-risk subgroups in the population and provide better insight into pathway mechanisms for complex diseases.
Current GWAS methods are designed to detect main effects, that is, direct associations of a single nucleotide polymorphism (SNP) or clusters of SNPs with disease.12, 13 In the context of complex diseases, examining main effects only could miss important genetic variants specific to subgroups of the population.
Gene-Gene Interaction Studies
Lichtenstein et al.14 studied twins and sought to connect hereditary factors to the causes of sporadic cancer. The study assessed the risks of cancer at 28 anatomical sites for twin children of a parent who has cancer. Statistical modeling was used to estimate the relative importance of heritable and environmental factors in causing cancer at 11 of those sites. A major finding was that inherited genetic factors make a minor contribution to susceptibility for most types of neoplasms, indicating that the environment plays the principal role in causing sporadic cancer. The relatively large effect of heritability in cancer at a few sites (such as prostate and colorectal cancer) suggests major gaps in our knowledge of the genetics of cancer.
Another large study, by Pearce et al.,15 that also focused on cancer attempted to link several well-established environmental risk factors for ovarian cancer and the results of a recent GWAS that identified six variants that influence disease risk. They pooled data from 14 ovarian cancer case-control studies, and then conducted stratified analyses of each environmental risk factor to evaluate the presence of interactions for all histological subtypes. They fit a multivariate model to examine the association between all environmental risk factors and genetic risk score on ovarian cancer risk. The results indicated no strong statistical evidence of interaction between the six SNPs or genetic risk score and the environmental risk factors on ovarian cancer risk.
A large bladder cancer study16 demonstrated interactions due to smoking using a logistic regression (LR) adjusted for age. This study coded the genotype variable as a count of minor alleles conforming to our Models 1, 2, and 3 below. A study involving prostate cancer17 found no contribution from a number of environmental factors. This study used a number of LR models similar to those we used in our analysis. Another study18 developed a Bayesian framework to investigate the influence of multiple loci simultaneously on disease risk. Their “full” model consisted of a standard LR model that treats the genotype variable as a categorical variable and specifies a main effect with interactions model.
Researchers have also used GWAS to examine type 2 diabetes, a second disease with a strong interplay of both environmental and genetic factors.19 Genetic loci discovered through GWAS explain only a small portion of the disease risk variance; some of the unexplained risk may be due to gene-environment interactions. The study suggested that the adverse effect of several type 2 diabetes loci may be abolished or at least attenuated by higher physical activity levels or healthy lifestyle, whereas low physical activity and the typical Western diet may augment it.
Patel et al.19 used data from two surveys from the Centers for Disease Control and Prevention’s National Health and Nutrition Examination Survey (NHANES). They used a GWAS to screen 18 genetic loci and type 2 diabetes for statistical interactions that were associated with disease. They describe their investigation as an environment-wide association study (EWAS), and they used data sets from four cohorts from the NHANES. Because the four cohorts were analyzed individually, the number of environmental factors varied among them.
Patel et al.19 used logistic regression and adjusted all models for age, sex, body mass index, and race. The results identified eight potential disease gene–environmental factor interactions. One interaction (trans-β-carotene) was particularly significant. The per-risk-allele effect sizes, after adjusting for age, sex, body mass index, and race for subjects with low trans-β-carotene levels, were 40 percent greater than the marginal genetic effect size of the SNP. They also found a strong interaction between a SNP and a nutrient found in corn oil, which conveyed a 20 percent higher risk than the SNP alone did.
Murcray et al.20 performed a general methodological study that focused on identifying SNPs that demonstrate heterogeneity between subgroups defined by some environmental exposure. They describe a two-step approach for detecting loci involved in gene-environment interactions that is performed independently of any initial scans for main effects. They expanded on the traditional test for gene-environment interaction in a case-control study by incorporating a preliminary screening step constructed to efficiently use all available information in the data. They claim that their two-step approach is more powerful than the standard test of interaction across a wide range of models and consequently is more robust to changes in environmental exposure and minor allele frequency than the traditional one-step test for identifying highly significant SNPs. The difficulty with most methods, including theirs, is that it is not a “data mining” method. The specific environmental factor and the form of that factor have to be established prior to analysis. This has proven to be a difficulty with our methods as well. The specific environmental factor or factors to include in the model greatly affect the power of the tests. Specifically, researchers should use some combination of the literature and/or data mining activities to establish the form of the environmental effect model (step function or linear) on the logistic scale.
A study by Cornelis et al.21 provides a comparative study of several logistic regression–based tests of gene-environment (G-E) and G×E interactions. All seven methods compared in their paper assumed a log-additive mode-of-inheritance model for each SNP. This differs from our methods, in which the mode of inheritance was agnostic. Cornelis et al. do not identify a preference for any of the seven methods and instead indicate that preference would depend on the goal of the study. They also explored methods investigating environment effects only in subjects with a positive phenotype case (i.e., case-only studies).
Finally, a Kraft et al. study,22 similar in content to the Cornelis et al. study in that it also focused on log-additive gene models, formulated a likelihood ratio test of association between disease and locus with the possibility that the genetic effect may be modified by an environmental factor. The specific environment model they investigated was similar to one of the experiments examined in our study—namely, a chemical spill—an all-or-nothing type of exposure.
Methods
Overview
We simulated genetic and environmental interactions in a GWAS context using a qualitative association framework to determine which statistical methods and models reliably predict associations between a qualitative phenotype (specifically, a disease diagnosis, coded as “case” for a positive diagnosis or “control” for a negative diagnosis) and a gene paired with an environmental influence. As with our previous work,2 the concept of relative risk is the basis for this investigation. We define the genetic relative risk (Φ) of a wild-type genotype to be the ratio of the probability of a positive diagnosis given an occurrence of a (wild-type) genotype divided by the probability of disease in the absence of the disease genotype. We also define the environmental risk (Π) as the ratio of the probability of a positive diagnosis given an exposure divided by the probability of a positive diagnosis in unexposed subjects. The values of Φ and Π are specified exogenously and vary from low-risk to not-so-low-risk.
We generated 1,000 replicates of simulation data that depended on the two risk values (Φ and Π) for each of three gene models using a standard Bernoulli process and analyzed them in terms of the observed power profiles for a low alpha error (α ≤ 10−8). The distribution of the number of alleles per genotype was randomized across replicates and was based on real data from Schymick et al. (2007).23 We biased the risk levels to the low end of the risk continuum because these are more difficult scenarios and are typical of what has been observed in the literature.7 To support these low risk levels, we fixed our sample size to N = 10,000 (5,000 cases and 5,000 controls) and N = 200,000 (100,000 cases and 100,000 controls) to determine whether it is possible to measure associations in low-risk, recessive inheritance scenarios. Others24 have used smaller values (N = 6000) for comparable investigations.
Generating the Synthetic SNP Data
We derived our data generation method from a study by Iles25 and from Mendelian concepts of inheritance. We specifically incorporated autosomal dominant, recessive, and additive inheritance patterns into the data. These data also depend on factors known to influence association measurements in the context of GWAS. Our simulation process assumes Mendelian type inheritance patterns.
Penetrance was defined as the proportion of individuals without the risk allele who have a definable trait (phenotype). In other words, penetrance was a genotype-specific probability of being affected with the trait. We designated a as the risk allele and A as the allele without risk. Generating the synthetic dataset using the relationships between penetrance and risk for different mode of inheritance (MOI) categories was straightforward (see Cooley et al., 2010,2 for further detail).
Initially, we supplied as input data the following variables:
- n = the target number of cases and controls in a given experiment,
- P = the disease penetrance,
- Φ = the genotype relative risk (1.10, 1.15, 1.20), and
- Π = the environmental relative risk (specification details are provided below).
The distribution of genotypes were drawn at random from a master set of genotype distributions obtained from real SNP data.23
In screening samples from the master set, Chan et al.26 recommend that a minor allele frequency (MAF) threshold not be applied as a filter. They argue that filtering MAFs out of the process because of low frequencies or to maintain Hardy–Weinberg equilibrium deviation has little effect on the overall false positive rate and, in some cases, filtering on MAF excludes SNPs. The effect of this step is to select a specific genotype distribution at random from the master distribution.
From the selected relative risk (Φ), penetrance (P), and MOI assumptions, we used the formulas in Table 1 to assign a case (1) or control code (0). This step converts the relative risk ratio (Φ) into the probability of a case (disease), given the MOI gene model assumed.
Table 1
Relative risk assumptions by mode of inheritance (MOI).
This genotype-specific process can be represented by the following logic:
- Major homozygote (AA)If the AA (non-disease) genotype is selected, the probability of a case equals the disease penetrance, P.
- Minor homozygote (aa)Ψ aa is the exogenous risk and represents the ratio of two probabilities: the probability of a case for a minor homozygote divided by the probability (Pr) of a case for a major homozygote, i.e.,Ψ aa = Pr(case/ aa)/Pr(case/ AA) = x /P.1Thus, the probability of a case given the minor genotype isx = Ψ aa * P2
- Heterozygote (aA)By the same argument, the phenotype risk given a heterozygote isΨ aA = Pr(case/ aA)/Pr(case/ AA) = y /P.3Thus, the risk of a case given the heterozygote genotype isy = Ψ aA * P4where Ψ aA is the assumed risk factor and P is the assumed penetrance.
Implicit in equations 1 through 4 is a consistent definition of penetrance defined as the proportion of cases that are present in the major genotype AA.
Using the estimate of x from equation 2 and y from equation 4, we specified a subject as a case (1) or control (0) at random using the four different MOI models from Table 1. For the MOI models that assume an elevated risk from the minor and the heterozygote genotypes, we would expect a higher proportion of cases to be more easily identified via the statistical procedures. Specifying risk depends on known and unknown disease mechanisms. A relative risk of 1.7 is considered strong and is associated with positive replication,27 and a risk of 1.3 is considered28 to be a more realistic assumption for complex diseases. Consequently, we limited our focus to relative risks in the range of 1.10 to 1.20.
Note that we assigned cases and controls so that there would be no possibility for the introduction of bias. We chose to ignore errors in both genotype and the phenotype measurements which in a real experiment could be a source of bias (we examined both sources of error in an earlier study).29 This process continued until we created n1 cases and n2 controls. We then applied a set of statistical methods (identified below) to predict associations, then recorded and tracked the results. For each set of unique factor combinations (i.e., penetrance, sample sizes, relative risk levels, and MOI categories) we generated 1,000 replicate experiments.
Exogenously, we specified the genetic inheritance (GI) relative risk of disease as 1.10, 1.15, and 1.20 and defined it in the Overview as the ratio of the probability of a disease diagnosis for subjects, dividing the wild-type gene by the probability of disease, based on all genetic and nongenetic causes. We also defined a second relative risk component based on a specific environmental exposure (EE). We defined this ratio as the probability of a disease given the environmental exposure divided by the probability of a diagnosis given no environmental exposure. In discussion of these experiments, we use the notation Φ to represent GI and Π to represent EE.
The form of the EE relative risk can be specified using a variety of assumptions. In all scenarios, the genetic risk is first used to determine the phenotype status (case or control). Then the environmental risk calculation determines whether the phenotype status is altered from control to case according to the EE assumptions. We assume that the form of the EE effect is not known but that the specific variable is known. In the following experiments we use E = age as a proxy for the different assumed forms of exposure, and we assign E a value obtained from a uniform distribution of 30 to 70. The value of E controls the EE risk according to different experiment designs. The main objective of this assessment is to identify whether one statistical model outperforms all other models and how much variation occurs across the different experiments.
For all experiments below, we used the GI as described above.
Experiment 1—The Main Effects Model
For the first experiment, half of the population (selected at random and assigned ages 50 < E < 71) incurred an EE relative risk (Π). The assigned risk value was 1.10, 1.20, 1.30, or 1.40. The other half of the population (assigned ages 29 < E < 51) incurred no risk; i.e., Π = 1.0. Thus, Experiment 1 simulates a fixed EE. When the determinant risk variable, E, exceeds a threshold, a positive diagnosis is more likely to occur. This is identified as the fixed risk, main effects, no interaction model.
Experiment 2—The Interaction Effects Model
For the second scenario, again half of the population (selected at random and assigned ages 50 < E < 71) incurred an EE relative risk (Π). This risk value was 1.10, 1.20, 1.30, or 1.40, but only if the subject also had a wild-type allele (i.e., a heterozygote or minor homozygote genotype). The other component of the population (aged 50 < E < 71 and genotype = AA) incurred no EE risk; i.e., Π = 1.0. Experiment 2 also simulates a fixed EE but only if the genotype contains a wild type allele. This is identified as the fixed risk, main effect with interaction model.
Experiment 3—The Main Effects Log-Linear Risk Model
For the third scenario, the entire population (randomly assigned ages 30 ≤ E ≤ 70) incurred an EE relative risk (Π) which was related to E in the following manner:
Experiment 3 simulates a log-linear variable risk model, with larger values of E conveying additional risk levels. As in Experiment 1, there is no interaction between the GI and EE risks.
Experiment 4—The Interaction Effects Log-Linear Risk Model
The fourth scenario is the same as the third scenario, but the risk applies only if the subject has a wild-type allele.
Experiment 4 simulates a variable risk scenario with larger values of E conveying higher risk levels—but only if the genotype contains a wild-type allele. This is the log-linear variable risk main effect with genotype interaction model.
For each experiment type, we varied the gene model to determine the relative power differences across model specification. Overall, Experiment 1 data has a step function relationship to EE and no interaction or difference in slopes (or EE step heights) across the three genotypes. In contrast, the Experiment 2 data has a step function relationship with EE where the aa and aA genotypes have the same slope (step height) but different intercepts. The AA genotype relationship to the EE is flat or has zero slope (no step up). In Experiment 3, the relationship to EE is log-linear, with equal slopes for all three genotypes. Finally, in Experiment 4, the relationship to EE is log-linear; the aa and aA genotypes have the same slope but different intercepts; and the AA genotype relationship to the EE is flat, or has zero slope.
Statistical Models
All models tested assumed a logistic regression (LR) specification. This form is commonly used in association studies involving environmental interactions.21
The variables used in the different models are shown in Table 2.
Table 2
Descriptions of variables used in the logistic regression models.
The difference between the experiments is straightforward. For subjects younger than age 50, the environmental exposure risk is 1.0 (i.e., no risk) in Experiments 1 and 2; subjects older than age 50 have an environmental exposure (i.e., the risk is greater than 1.0). However, for Experiment 2, an additional condition pertains: Here only subjects age 50 and older who have a wild-type allele are assumed to have the assigned risk. The main discriminator between Experiments 1 and 3 (and Experiments 2 and 4) is the risk characterization. For Experiments 1 and 2, the risk is intended to be an all-or-nothing process akin to a toxic exposure that occurs some time after the subject reaches age 50. For Experiments 3 and 4, the risk due to an environmental exposure is present in all subjects and increases as age increases. The experiments are summarized in Table 3.
Table 3
Experiment description.
We used three specific statistical models to assess the data generated by the four experiments. Each assumed an intercept term and had the following form:
- Model 1 is a logistic regression model with a single variable genotype (G) main effect (2 df). This is a candidate model if no environmental exposure were suspected.
- Model 2 is a logistic regression mixed main effects and interaction model (g1, g2, E, g1*E, g2*E) (6df). This is a fully specified model that assumes that the environmental exposure is a continuous variable.
- Model 3 is also a logistic regression mixed main effects and interaction model (g1, g2, e1, g1*e1, g2*e1) (6 df). This is the fully specified categorical model and assumes that the environmental exposure has a specific (all-or-nothing) categorical variable form.
The specific regression models we used in this study are summarized in Table 4 . Note that we initially compared six models. Two were gene-only models—a 1 df (log-additive test) and a 2 df test—and four were main effects plus interaction models. We had two environmental exposure specifications (E and e1) and two genetic inheritance specifications (G and g1, g2). From the six initial models, we selected the three models that dominated the other three: M-1, M-2, and M-3. We dropped the other three models (M-1a, M-2a, and M-3a) from our assessment.
Table 4
Statistical models assessed.
The test statistics we used in our analyses are defined as the difference between two log-likelihood (LLH) statistics. The first is specific to the model used, and the second is based on a model with only the intercept term.
Results
Association Analysis
In this section, we describe the power profiles that result by applying the models described in Table 4 to the data generated according to the four different experiments in Table 3. We focus on detecting the associations between the combined genotype-environmental factors on phenotype outcome (disease diagnosis). We assess the importance of model specification in predicting the presence of association with a phenotype of interest and to what degree the gene model and genotype environment interactions influence power. In the following section, Genotype Associations (p. 14), we assess the role of the genotype alone in predicting association while controlling for the environmental influence.
Note that in calculating all power results in this section we assumed that the Type I error rate was 10 −8 .
However, since all combined environmental exposure and genetic inheritance risk values are greater than 1.0 in all of our experiments, only Type II errors were possible.
Table 5 shows the data generated using the protocol for Experiment 1. Note that for this and all subsequent tables in this section, the highest power value for each risk profile within the three MOI categories is bolded to highlight the optimal model. For each genetic inheritance (GI) risk level (Φ) there is an environmental exposure (EE)risk level (Π) equal to 1.0, indicating no EE risk.
Table 5
Power values, by statistical model, Φ, and Π: Experiment 1—all gene models.
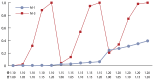
Figure 1 shows the data generated using the protocol for Experiment 1 for the additive gene model. Figure 1 includes the optimal model (Model M-3, identified by the bolded cells in Table 5) and the model that does not include an EE variable in its specification (Model M-1). The results presented in Table 5 and Figure 1 indicate that there is little difference in performance between models when the risk of EE is not present.
The results shown in Figure 1 and Table 5 indicate the following:
- The power profile of model M-1 is substantially below that of models M-2 and M-3. M-1 represents a typical single locus method used in a GWAS that ignores environmental influences. We conclude that not including an EE reduces the likelihood of the locus being associated with the phenotype.
- Model M-3 is the most powerful of the three models. This is expected since the Experiment 1 protocol should generate data consistent with the M-3 model formulation.
- The difference between the profiles of models M-2 and M-3 is a result of the manner used to characterize the EE functional form. Because the data was generated in a manner compatible with the e1 variable used in model M-3, it generated more accurate power predictions.
Note that in the full M-3 model, the overall intercept is the log of the intercept for the line that relates EE to the log-odds risk among those subjects with the non-disease genotype AA. The coefficient associated with the g1 main effect is testing for the difference between intercepts for the subjects with genotype aA and those with genotype AA. Similarly, the g2 main effect coefficient is testing for the difference between the intercepts for subjects with the aa genotype and those with the AA genotype.
The EE main effect coefficient is the height of the step in the step function relating EE to log-odds-risk for subjects with the AA genotype and, it therefore, tests for a common EE step height across all three genotypes. The g1*E interaction coefficient is the difference between the step heights for the aA subjects and the AA subjects. Similarly, the g2*E coefficient is the difference between the step heights for the aa subjects and the AA subjects. Since the AA, aA, and aa step heights/slopes associated with the EE environmental effect are all equal in Experiments 1 and 3, only the common main effect (ME) associated with EE contributes to association prediction in those data sets, and the interaction terms are superfluous.
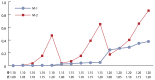
Table 6 and Figure 2 show the results of applying the three models described in Table 4 to the data generated according to the Experiment 2 protocol (see Table 3). Experiment 2’s results indicate that
- Even though model M-1 does not adjust for EE, the observed (relatively) high power profiles for high EE risk levels suggest that the GI-EE interaction effect is embedded in the M-1 power values, and the high power profiles are credited as a genotype main effect.
- As in Experiment 1, model M-3 outperforms all other models because the variable e1 properly characterizes EE behavior. This clearly demonstrates the value of preprocessing (i.e., mining) the data before committing to a specific association model.
Table 6
Power values, by statistical model, Φ, and Π: Experiment 2—all gene models.
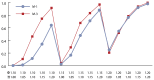
Table 7 and Figure 3 show the results of applying the three statistical models described in Table 4 to the data generated according to the Experiment 3 protocol (see Table 3 ).
Table 7
Power values, by statistical model, Φ, and Π: Experiment 3—all gene models.
For Experiment 3, the results shown in Figure 3 and Table 7 indicate that
- Model M-1 consistently performs below M-2 and M-3, indicating that not including an EE term limits the association assessment.
- In general, model M-2 produces better power profiles than M-3. This is expected given that the EE incremental risk is linearly related to the log of EE. Thus, model M-2 is more consistent with the protocol used to generate the data in Experiment 3.
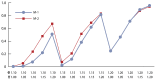
The results for Experiment 4 are shown in Table 8 and Figure 4. They indicate the results of applying the three models described in Table 4 to the data generated according to the Experiment 4 protocol (see Table 3).
Table 8
Power values, by statistical model, Φ, and Π: Experiment 4—all gene models.
The results shown in Table 8 and Figure 4 indicate that
- Consistent with Experiment 2’s results, model M-1 does not adjust for EE, but because of the influence of GI-EE interaction effects, M-1 displays higher power profiles for large EE risk levels.
- As in Experiment 3, model M-2 outperforms M-3 because it better characterizes the EE by using the variable E (age) and further demonstrates the value of preprocessing (i.e., mining) the data before committing to a specific association model.
- In the presence of GI-EE interaction effects, the genetic-only model (M-1) performs better than anticipated.
Genotype Associations
The analysis in the previous section focused exclusively on composite associations, that is, whether a specific gene plus an environmental factor associates with a phenotype. As we noted earlier, our main interest was separating main genetic effects from environmental effects and their interactions. To accomplish this, we defined a total effect test (TOT) that adjusts for EE where:
is the test we applied to the data generated by Experiments 1 and 2 protocols and
is the test that we applied to the data generated by Experiments 3 and 4 protocols.
TOT is the association test that measures genetic effects (main and interactive) and is adjusted for the environmental effect.30 TOT simultaneously measures whether the aA and aa intercepts are different from the AA intercept and whether the aA and aa slopes are non-zero, given that the AA slope on EE is zero. This test was used to test for association from all causes.
We also define two additional tests for genotype-environment interactions, INT, as follows:
and
The INT test subtracts the main effects for g1, g2, and EE from the TOT and tests whether the EE steps (or slopes) for the aA and aa genotypes are different from the corresponding EE step (slope) for genotype AA.
The final test measures the influence of the genetic main effects (ME).
is the test applied to the data generated by Experiments 1 and 2 protocols and
is the corresponding test for data from Experiments 3 and 4 protocols.
The ME tests check whether the estimated aA and aa intercepts differ from the AA intercept, conditioned on the EE step sizes (e1 in experiments 1 and 2) or the EE slopes (E in experiments 3 and 4) being equal for all three genotypes.
Note that for Experiments 2 and 4, both the AA step (coefficient of e1) and slope (coefficient of E) on EE are zero, and therefore the coefficient for the EE main effect (assuming that the M-3 is operating) is estimating zero; the two interaction columns are estimating the aA step/slope minus zero and the aa step/slope minus zero, respectively.
Typically, these three tests would be applied sequentially: TOT followed by INT, then ME. Assessing whether an interactive or non-interactive genetic association is obtained would depend on the result of the preceding test.
For example, if TOT is non-significant, the process stops and we conclude that there is no connection between the genetic locus and the phenotype. Otherwise, we would apply the INT test. If INT was significant, we could conclude that the locus and the phenotype are significantly related, with the caveat that the strength of the genotype effect varies by the EE risk level. The ME test would only be applied if the TOT is significant and INT test is not significant. In this case, the ME test would be applied to affirm that the genetic and environmental effects are operating independently of each other and to assert that a common genotype main effect exists that applies to all EE levels. The results of running the three tests (TOT, INT, and ME) are shown in Tables 9 through 13.
Table 9
Total effects test (TOT) power values, by risk profile, Φ, and Π—all experiments and gene models,N= 200,000.
Table 10
Genotype-environment interactions (INT) power values, by risk profile (Φ and Π)—all experiments and gene models,N= 10,000.
Table 11
Main effects (ME) power values, by risk profile (Φ and Π)—all gene models,N = 10,000.
Table 12
Genotype-environment interactions (INT) power values, by risk profile (Φ and Π)—all gene models,N= 200,000.
Table 13
Main effects (ME) power values, by risk profile (Φ and Π)—all gene models,N = 200,0000.
Consider that every replicate in every cell produced by the simulation experiments is designed to generate a genotype-phenotype association (albeit at low risk). Some of these replicates influenced by an EE also contribute toward association. However, in a perfect statistical world, all are generated to predict an association with the phenotype. The fact that they do not is an indication of the limitations of the GWAS process.
In addition, Table 9 suggests that
- Association detection involving recessive genes is difficult to identify (and accordingly requires a larger sample size than we used in our experiments.
- Scenarios involving gene-environment interactions (Experiments 2 and 4) greatly influence whether genetic influences can be detected by a gene-only model.
- The type of EE process influences the ability to detect an association, whether the effect is due to a chemical-type exposure (Experiments 1 and 2) or is due to aging (Experiments 3 and 4).
Table 10 presents the results of applying the INT test to all experiments and all gene models. Not shown are the results for Experiments 1 and 3, which generated data without interaction effects. They estimate no interaction between GI and EE (as they should), so those results are not shown. Note that the Type 1 α thresholds in Table 11 for generating power estimates for all cells are ≤10 −2.
Table 10 reaffirms the results of Table 9, namely, that
- Power values for recessive genes are very low and accordingly were more difficult to identify than other gene models.
- Gene-environment interactions influence association outcomes. This is evidenced by all cells of the no-interaction experiments (1 and 3) having power values <.004.
- The type of EE process influences the detection of an association, whether the effect is due to an exposure (Experiments 1 and 2) or is due to an aging mechanism (Experiments 3 and 4).
- Interaction effects achieve significant levels in Experiment 2 for risk values of EE ≥ 1.2 only.
Table 11 presents the results of the ME test for Experiments 1 and 3. ME results for Experiments 2 and 4 are not shown because they were generated by a protocol that produced EE and GI interactions, and if the INT test demonstrated significance (as it should have), the ME tests would have been unnecessary. In all cases the alpha threshold was set to 10−2.
Table 11 reaffirms the results of Tables 9 and 10: namely, that
- Associations involving recessive genes are more difficult to identify,
- Gene-environment interactions influence association outcomes,
- The type of process influences the detection of an association, as shown by differences between power values for exposure mechanisms such as those resembling chemical spills (Experiments 1 and 2) and those recognizing aging mechanisms (Experiments 3 and 4), and
- Main effects are only ascribed significant for larger risk values of genetic inheritance, those with a risk of 1.2 or above.
Note that the power threshold values are set to low (α ≤ 10−2) for the interaction and main effects tables. To investigate the effect of a very large N, we repeated the simulation process with N = 200,000 and reduced the threshold to (α ≤ 10−8 ). The results are shown in Tables 12 and 13.
The results shown in Table 12 suggest that the GI-EE interactions are very sensitive to low EE levels (Π < 1.10). They also accurately estimate an interaction power value of zero when Π = 1.00, that is, no EE risk.
These results suggest that for very large studies it is possible to predict positive associations between recessive genes linked to phenotypes with low to moderate risk.
Conclusions
In summary, the chances of predicting an association in a genome-wide association study are reduced if an environmental effect is present and the statistical model does not adjust for it. This is especially true if the environmental effect and genetic marker do not have an interaction effect. The functional form of the model also matters. The more accurately the form of the environmental influence is portrayed by the statistical model, the more accurate the prediction will be. Even with very large sample sizes, association predictions involving recessive markers are low.
This study focused on one important methodological step involved in conducting a GWAS: selecting a statistical method and a supporting model that reliably predict associations. This study does not address the broader issue of the supporting experimental design that employs the statistical methods as part of an overall solution strategy. Those combined issues and their mutual interconnections are described by Cordell.31
The specific scenarios we address here involve genetic associations that have environmental influences. Our assumption is that the environmental influence that contributes to a given phenotype is in question and the precise form of that influence is unknown. A separate analysis to characterize the functional form to proxy the mechanism behind the environmental exposure is required. These approaches should focus on case-only data similar to the methods described in the Cornelis et al. study.21 These approaches involve investigating different environmentally related functional relationships between the suspected environmental influence and the phenotype in the cases-only subpopulation. For example, if gene effects and environmental effects are independently significant with respect to disease prevalence, a polynomial model could be used to characterize the relationship between environmental effects and the log-odds of disease prevalence. This would allow testing whether the nonlinear parameterization was required to characterize the environmental effect. Alternatively, if the environmental effect has multiple levels such as age, researchers could investigate a cubic polynomial to assess whether the effect stayed low initially then rose at some point and flattened out toward the end of the environmental effects range. If this analysis suggests an appropriate polynomial level for environmental effects, researchers should also investigate a similar assessment using the gene-environment interaction variable.
We have used this simulation scenario in previous studies. We reviewed single gene models and evaluated a wide class of statistical methods.2 Our results indicated that researchers should consider a multi-test procedure that combines individual gene-based (dominant, recessive, additive) core tests as a composite statistical method for conducting the initial screen in a GWAS. The tests can be combined into a single operational test in a number of ways. Two such tests are the Bonferroni procedure32 and the MAX procedure,33 which produce very similar statistical power profiles. Of course, if the gene model under investigation is known, a single test that assumes the implied form is better than a combined test. However, for this study all patterns across gene models are consistent and only vary by degree.
Elsewhere, we have also evaluated the effect of phenotype errors that resulted from inaccurate diagnoses and genotype errors that resulted from gene-chip errors or occurrences of DNA methylation altering gene expression that associate a wild-type gene with the wrong phenotype outcome.29 Our results quantify the relationship between genotype and diagnosis error measures and sample size to achieve a .80 statistical power level. Our results also demonstrate that researchers should not underestimate the need to increase sample size to compensate for power loss due to the presence of genotype and diagnosis errors.
We also investigated epistatic scenarios involving two genes.34 The results showed that the most powerful statistical methods for predicting associations between phenotypes and genotypes in epistatic scenarios are statistical models that simultaneously test for associations involving both interacting loci. This is consistent with the results we present here. This result is not surprising and has been reported by others. We reported that if two genes contribute to a phenotype, the weaker gene will be obscured by the stronger gene and often not be identified as a contributor to the phenotype when a single gene model is used. Again, this result is similar to showing that the effect of an environmental exposure can obscure the influence of a genotype-phenotype association if the model does not account for the GI and the EE simultaneously. In this sense, two-gene models (or alternatively a gene-environment model) produce better predictions of association than single-gene models do.
We acknowledge that our results could possibly depend on the particular experiments we devised to investigate how the statistical models performed. In light of this, we are reviewing other scenarios to establish the robustness of our findings. Nevertheless, establishing the genotype-to-phenotype connections without using a simulation approach is limited.
For the gene-environment interaction scenarios addressed here, the results across all gene models lead us to conclude that using a composite test that supports distinct underlying statistical models, that is, a “main effects–only” model and a “main effects with interactions” model, is likely to be more effective than single model tests; this result does not depend on the gene model and thus differs from the single gene and epistatic scenarios, where each different gene model assumption (i.e., recessive, dominant, and additive) requires representation in the composite test.2, 32
References
- 1.
- Kuo C-L, Feingold E. What’s the best statistic for a simple test of genetic association in a case-control study? Genet Epidemiol. 2010; 34(3): 246 –53. [PubMed: 20025064]
- 2.
- Cooley P, Clark R, Folsom R, Page G. Genetic inheritance and genome wide association statistical test performance. J Proteomics Bioinform. 2010; 3: 321–5.
- 3.
- Li J, Horstman B, Chen Y. Detecting epistatic effects in association studies at a genomic level based on an ensemble approach . Bioinformatics 2011 Jul 1; 27(13): i222–9. [PMC free article: PMC3117367] [PubMed: 21685074]
- 4.
- Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004 May 27; 429(6990): 446–52. [PubMed: 15164069]
- 5.
- Manolio TA. Genome-wide association studies and assessment of the risk of disease. N Engl J Med. 2010 Jul 8; 363(2): 166–76. [PubMed: 20647212]
- 6.
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005 Apr; 37(4): 413–7. [PubMed: 15793588]
- 7.
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011 Sep 1; 477(7362): 54–60. [PMC free article: PMC3832838] [PubMed: 21886157]
- 8.
- Spencer C, Hechter E, Vukcevic D, Donnelly P. Quantifying the underestimation of relative risks from genome-wide association studies. PLoS Genet. 2011 Mar; 7(3): e1001337. [PMC free article: PMC3060077] [PubMed: 21437273]
- 9.
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009 Oct 8; 461(7265): 747–53. [PMC free article: PMC2831613] [PubMed: 19812666]
- 10.
- Terry PD, Umbach DM, Taylor JA. APE1 genotype and risk of bladder cancer: evidence for effect modification by smoking. Int J Cancer. 2006 Jun 15; 118(12): 3170–3. [PubMed: 16425270]
- 11.
- Stern MC, Johnson LR, Bell DA, Taylor JA. XPD codon 751 polymorphism, metabolism genes, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002 Oct; 11(10 Pt 1): 1004–11. [PubMed: 12376500]
- 12.
- Browning BL, Browning SR. Efficient multilocus association testing for whole genome association studies using localized haplotype clustering. Genet Epidemiol. 2007 Jul; 31(5): 365–75. [PubMed: 17326099]
- 13.
- Zhao J, Jin L, Xiong M. Nonlinear tests for genome wide association studies. Genetics. 2006 Nov; 174(3): 1529–38. [PMC free article: PMC1667094] [PubMed: 16816420]
- 14.
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer— analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000 Jul 13; 343(2): 78–85. [PubMed: 10891514]
- 15.
- Pearce CL, Rossing MA, Lee AW, Ness RB, Webb PM, Chenevix-Trench G, et al. Combined and interactive effects of environmental and GWAS-identified risk factors in ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2013 May; 22(5): 880–90. [PMC free article: PMC3963289] [PubMed: 23462924]
- 16.
- Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nature Genet. 2010 Nov; 42(11): 978–84. [PMC free article: PMC3049891] [PubMed: 20972438]
- 17.
- Lindstrom S, Schumacher F, Siddiq A, Travis RC, Campa D, Berndt SI, et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers— results from BPC3. PloS One. 2011; 6(2): e17142. [PMC free article: PMC3044744] [PubMed: 21390317]
- 18.
- Yu K, Wacholder S, Wheeler W, Wang Z, Caporaso N, Landi MT, et al. A flexible Bayesian model for studying gene-environment interaction. PLoS Genet. 2012 Jan; 8(1): e1002482. [PMC free article: PMC3266891] [PubMed: 22291610]
- 19.
- Patel CJ, Chen R, Kodama K, Ioannidis JP, Butte AJ. Systematic identification of interaction effects between genome- and environment-wide associations in type 2 diabetes mellitus. Hum Genet. 2013 May; 132(5): 495–508. [PMC free article: PMC3625410] [PubMed: 23334806]
- 20.
- Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009 Jan 15; 169(2): 219–26. [PMC free article: PMC2732981] [PubMed: 19022827]
- 21.
- Cornelis MC, Tchetgen EJ, Liang L, Qi L, Chatterjee N, Hu FB, et al. Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. Am J Epidemiol. 2012 Feb 1; 175(3): 191–202. [PMC free article: PMC3261439] [PubMed: 22199026]
- 22.
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007; 63:(2) 111 –9. [PubMed: 17283440]
- 23.
- Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2007 Apr; 6(4): 322–8. [PubMed: 17362836]
- 24.
- Thornton KR, Foran AJ, Long AD. Properties and modeling of GWAS when complex disease risk is due to non-complementing, deleterious mutations in genes of large effect. PLoS Genet. 2013; 9(2): e1003258. [PMC free article: PMC3578756] [PubMed: 23437004]
- 25.
- Iles MM. Effect of mode of inheritance when calculating the power of a transmission/disequilibrium test study. Hum Hered. 2002; 53:(3) 153–7. [PubMed: 12145551]
- 26.
- Chan EK, Hawken R, Reverter A. The combined effect of SNP-marker and phenotype attributes in genome-wide association studies. Anim Genet. 2009 Apr; 40(2): 149–56. [PMC free article: PMC2680326] [PubMed: 19076733]
- 27.
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007 Feb 22; 445(7130): 881–5. [PubMed: 17293876]
- 28.
- Ziegler A, Konig IR, Thompson JR. Biostatistical aspects of genome-wide association studies. Biom J. 2008 Feb; 50(1): 8–28. [PubMed: 18217698]
- 29.
- Cooley P, Clark RF, Page G. The influence of errors inherent in genome wide association studies (GWAS) in relation to single gene models. J Proteomics Bioinform. 2011 Jul; 4: 138–44.
- 30.
- Lehmann EL, Romano JP. Testing statistical hypotheses. New York: Springer; 2005.
- 31.
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009 Jun; 10(6): 392–404. [PMC free article: PMC2872761] [PubMed: 19434077]
- 32.
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979; 6:(2) 65–70.
- 33.
- Li Q, Zheng G, Li Z, Yu K. Efficient approximation of P-value of the maximum of correlated tests, with applications to genome-wide association studies. Ann Hum Genet 2008 May; 72(Pt 3): 397–406. [PubMed: 18318785]
- 34.
- Cooley P, Gaddis N, Folsom R, Wagener D. Conducting genome-wide association studies: epistasis scenarios. J Proteomics Bioinform 2012 Sep; 5(10): 245–51.
Philip C. Cooley, MS, is an RTI Senior Fellow in Bioinformatics and High-Performance Computing. His main interests are computational biology and infectious disease modeling. He has developed models to assess intervention strategies to contain pandemic influenza, tuberculosis, malaria, HIV/AIDS, and obesity.
Robert F. Clark, PhD, is a senior genetic epidemiologist in RTI International’s Genetic Epidemiology and Omics research program. Throughout most of his career, he has focused on multidisciplinary work in the omics and systems biology of various multifactorial disorders, and since 1992, he has conducted many genetic studies of neurodegenerative diseases; breast, bone, and brain cancers; nicotine, heroin, and cocaine addiction; and aortic aneurysms.
Ralph E. Folsom, PhD, is a chief scientist in RTI’s Center for Statistical and Data Sciences. He is an expert in designing and analyzing complex probability samples. Dr. Folsom leads RTI’s innovative work in small area estimation (SAE). He directs the National Survey on Drug Use and Health (NSDUH) project team that develops annual and biannual SAEs for drug use, dependency, treatment, and treatment need for states and sub-state regions.
RTI International is an independent, nonprofit research organization dedicated to improving the human condition. The RTI Press mission is to disseminate information about RTI research, analytic tools, and technical expertise to a national and international audience. RTI Press publications are peer-reviewed by at least two independent substantive experts and one or more Press editors.
Suggested Citation
Cooley, P.C., Clark, R.F., and Folsom, R.E. (2014). Assessing gene-environment interactions in genome wide association studies: statistical approaches. RTI Press Publication No. RR-0022-1405. Research Triangle Park, NC: RTI Press. https://doi.org/10.3768/rtipress.2014.rr.0022.1405
RTI International is an independent, nonprofit research institute dedicated to improving the human condition. We combine scientific rigor and technical expertise in social and laboratory sciences, engineering, and international development to deliver solutions to the critical needs of clients worldwide.
- NLM CatalogRelated NLM Catalog Entries
- PMCPubMed Central citations
- PubMedLinks to PubMed
- How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures?[Pac Symp Biocomput. 2018]How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures?Veturi Y, Ritchie MD. Pac Symp Biocomput. 2018; 23:228-239.
- Methodological and conceptual issues regarding occupational psychosocial coronary heart disease epidemiology.[Scand J Work Environ Health. 2...]Methodological and conceptual issues regarding occupational psychosocial coronary heart disease epidemiology.Burr H, Formazin M, Pohrt A. Scand J Work Environ Health. 2016 May 1; 42(3):251-5. Epub 2016 Mar 9.
- Part 1. Statistical Learning Methods for the Effects of Multiple Air Pollution Constituents.[Res Rep Health Eff Inst. 2015]Part 1. Statistical Learning Methods for the Effects of Multiple Air Pollution Constituents.Coull BA, Bobb JF, Wellenius GA, Kioumourtzoglou MA, Mittleman MA, Koutrakis P, Godleski JJ. Res Rep Health Eff Inst. 2015 Jun; (183 Pt 1-2):5-50.
- Review Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.[Brain Neurotrauma: Molecular, ...]Review Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.Wolahan SM, Hirt D, Glenn TC. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. 2015
- Review Applications of Monte Carlo Simulation in Modelling of Biochemical Processes.[Applications of Monte Carlo Me...]Review Applications of Monte Carlo Simulation in Modelling of Biochemical Processes.Tenekedjiev KI, Nikolova ND, Kolev K. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science. 2011 Feb 28
- Assessing Gene-Environment Interactions in Genome-Wide Association Studies: Stat...Assessing Gene-Environment Interactions in Genome-Wide Association Studies: Statistical Approaches
Your browsing activity is empty.
Activity recording is turned off.
See more...