This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. To view a copy of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Gilmore MS, Clewell DB, Ike Y, et al., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014-.

Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet].
Show detailsIntroduction
Enterococci are a versatile group of bacteria found in various habitats, which range from a commensal presence within the gastrointestinal tract of mammals and other organisms, to the environment where they have been identified within soil, water, and food supplies. They are also remarkably hardy and are able to withstand drastic changes within their environments, including factors of temperature, salinity, pH, and available nutrients (Klare, Werner, & Witte, 2001). The ability of the enterococci to rapidly respond to growth and environmental conditions is largely achieved by controlling gene expression. In this chapter, we focus on work to date that provides current understanding of the gene regulation mechanisms in Enterococcus faecalis, an opportunist pathogen with intrinsic resistance to many antibiotics and a causative agent of nosocomial urinary tract infections, endocarditis, and bacteremia.
Gene regulation in E. faecalis occurs on multiple levels. At the transcriptional level, the initiation of gene transcription is regulated by transcriptional factors which modulate promoter activities. Thanks to advancements in genome sequencing, multiple genomes of enterococcal species have been revealed, which provides the foundation for an understanding of enterococcal gene function and regulation processes. In the case of E. faecalis V583, the genome consists of 3337 open reading frames (ORFs) on the chromosome, as well as three plasmids (Paulsen, et al., 2003). Among these, 145 genes are predicted to encode for transcriptional regulators that belong to different protein families, including Cro/CI, GntR, MerR, and other transcription regulator protein families (Paulsen, et al., 2003). These transcriptional factors affect the expression of their effector genes by binding to the promoter region of these genes and positively or negatively affecting transcription, which subsequently alters the abundance of the specific mRNAs and proteins produced. This chapter discusses the mode of action for several transcriptional factors identified in E. faecalis and the genes that are regulated by them.
Another broad set of regulation machinery involved in gene expression are the two component systems that sense extracellular signaling molecules, and regulate target gene expression at the transcription level as a result. A typical two-component system consists of a membrane-associated histidine kinase receptor (sensor HPK) and a cognate response regulator (RR) which functions as a transcriptional factor (Stock, Robinson, & Goudreau, 200). Phosphorylation of the response regulator by the histidine kinase upon specific environmental stimuli alters its ability to bind to the target DNA sequence or interact with other components of the transcription machinery, which in turn regulates gene expression. In E. faecalis V583, at least 15 two-component systems have been identified that play a critical role in E. faecalis’ ability to response to a wide variety of stimuli, including quorum signals, antimicrobials, nutrients, serum components, and bile salts (Paulsen, et al., 2003). The mode of function of several two-component systems, including quorum-sensing related Fsr and vancomycin-resistant gene regulators VanRS, are further discussed in detail to demonstrate the ways in which bacteria respond to different environmental stimuli through these signal transduction processes.
Other than the typical HPK-RR two-component systems, several other signaling pathways involving transcriptional regulation in E. faecalis have been described, including the sex pheromone signaling pathway and the pathway for cytolysin toxin expression. In both of these systems, expression of the target genes are controlled by the balance of extracellular antagonistic peptide pairs, which are shifted upon the presence of recipient cells or host cells, respectively.
Besides transcriptional regulation, control of gene expression can also occur post-transcriptionally through other mechanisms, including mRNA processing, regulation via riboswitches, or modulation by antisense and small regulatory RNA (sRNA). One of the best-studied examples of post-transcriptional regulation is seen in the control of genes involved in ethanolamine utilization (Garsin, 2010). The E. faecalis ethanolamine utilization (eut) genes are encoded in a complex locus of 18 genes, and include structural, enzymatic, and regulatory components. Upregulation of the eut genes is triggered by the presence of two environmental cues—ethanolamine and adenosylcobalamin (AdoCbl). Each of these input signals is perceived and relayed by two different regulatory systems, and synchrony between them is required for expression of the eut genes. Both regulatory systems control gene expression at the post-transcription initiation level, and target nascent RNA instead of DNA. Post-transcriptional regulation also occurs at several levels in the control of conjugation of a pheromone response plasmid as previously discussed, in which antisense RNA plays an important role in transcriptional regulation. This RNA-mediated reciprocal regulation is RNAse III dependent, which demonstrates the importance of RNA processing in gene expression regulation. A similar observation is reported for the gene expression of E. faecalis Ebp pili, which is positively regulated by the novel endo- and exonuclease RNase J2 (Gao, Pinkston, Nallapareddy, van Hoof, Murray, & Harey, 2010). Small regulatory RNAs have recently been identified as important regulatory elements in bacterial gene expression. Recent transcriptomic analysis of E. faecalis sRNA provides valuable information to understand gene regulation through sRNAs. One of the newly identified sRNAs, EF3314_EF3, is potentially involved in the turnover of some abundant proteins, as demonstrated by the ΔEF3314_EF3315 sRNA mutant, a finding that indicates a novel regulation process (Shioya, et al., 2011).
Virulence Related Gene Expression Regulation
Known E. faecalis virulence factors include peptidoglycan-anchored surface proteins, such as Ace and the Ebp pili; secreted peptides, such as cytolysin; membrane bound proteins, including aggregation substance; extracellular enzymes, including gelatinase and serine Protease; and other gene products, which play important roles in response to different biological needs that range from surviving environmental changes or evading host defense to adherence and colonization. Similar to other bacterial pathogens, E. faecalis has developed sophisticated regulation machinery to tightly control the expression of virulence-related genes to its benefit. Understanding the regulation events in virulence factor expression will contribute significant value in the development of future strategies to battle enterococcal infections.
Transcriptional regulators: SlyA as an example
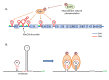
Proteins of the MarR/SlyA family are known to regulate the expression of virulence genes in many Gram-positive and Gram-negative organisms. Evidence of such a regulator existing in enterococci arose with the examination of the crystal structure of an E. faecalis transcriptional factor (EF3002) in 2003 (Wu, et al., 2003). The 150 amino acid SlyA protein is encoded by an open reading frame that begins with a GTG initiator codon. The crystal structure of SlyA was determined without its DNA target or signal molecule bound, and the unliganded crystal structure revealed that SlyA consists of two monomers in an asymmetric unit (Wu, et al., 2003) (Figure 1). In the SlyA crystal, key arginine residues thought to play a role in DNA binding were found to point away from the major groove, as was determined for the crystal structure of MarR (Alekshun, Levy, Mealy, Seaton, & Head, 2001), but differs from that of the MexR (Lim, Poole, & Strynadka, 2002) transcriptional regulator of Pseudomonas aeruginosa. The high degree of flexibility observed between the crystal structures of the various DNA-binding domains provides indirect evidence of the ability of this fold to adapt in order to recognize various DNA targets. Conserved residues among the various transcriptional regulator structures suggest that this fold could make contact with the phosphate groups or bases. Comparison of the SlyA-Ef structure with the structure of MarR and MexR suggest they are similar, which indicates that the MarR/SlyA family shares a common fold and similar DNA-binding properties, despite their low amino acid sequence similarity (Wu, et al., 2003) (Figure 1).
The promoter region of SlyA suggests that it is a part of a bicistronic operon that also serves as the promoter for EF3001, which encodes an N1-acetyltransferase enzyme. This transcriptional organization is conserved in other Gram positive species (Michaux, et al., 2011). It is thought that over-expression of EF3001 would be an advantage to the ΔslyA mutant as it shows homology to PaiA, a protein from Bacillus subtilis, that encodes for an N1-spermidine acetyltransferase (Forouhar, et al., 2005) involved in polyamine homeostasis (excess of this molecule is toxic to the cell).
Recent functional analyses of the SlyA regulator (ef3002) (Michaux, et al., 2011; Michaux, Martini, Hanin, Auffray, Hartke, & Giard, 2011) have illustrated that SlyA activity is part of a complex regulatory network. Observations have suggested that the involvement of SlyA is linked to virulence and persistence inside the host (Michaux, et al., 2011). SlyA mutant strains were found to be more virulent than the parental wild type strain in a wax moth (Galleria mellonella) model (Michaux, et al., 2011). The immune response of G. mellonella has a number of structural and functional similarities to the innate immune response of mammals (reviewed in (Vogel, Altincicek, Glöckner, & Vilcinskas, 2011)). The ΔslyA mutant was also found to survive better in mouse organs and in macrophages (Michaux, et al., 2011). DNA microarray experiments revealed that 117 genes were deregulated in the ΔslyA mutant as compared to the parental strain, a finding that suggests that SlyA acts as a repressor and activator (Michaux, et al., 2011)—although it should be noted that the expression of most of these genes changed by only two fold. EF3001 was the only gene in this analysis where expression in the ΔslyA mutant was up-regulated by 16.62-fold as compared to the parental wild type strain, suggesting that the enzyme it encodes may provide an important advantage during infection (Michaux, et al., 2011). Another gene encoding a helicase (EF3217) in the ΔslyA mutant was strongly down-regulated, but its role in virulence is unclear. This particular gene is associated with a mobile element and is not conserved across a number of the sequenced enterococcal genomes (Michaux, et al., 2011).
Further work using a pVEPhoZ-PslyA strain revealed that the expression level of the slyA operon was increased in the presence of bile salts (Michaux, Martini, Hanin, Auffray, Hartke, & Giard, 2011). Under the stressed conditions induced by the presence of 0.08% bile salts, the growth of the ΔslyA mutant was impaired, with the doubling time of the strain decreased by a factor of two. Under these growth conditions, the bile salt hydrolase (BSH) encoded by EF3005 in the E. faecalis V583 genome was found to be induced in the ΔslyA mutant by six-fold in comparison to the wild type parental strain, which is somewhat puzzling, given the growth phenotype of the ΔslyA mutant in the presence of bile salts (Michaux, Martini, Hanin, Auffray, Hartke, & Giard, 2011). In the E. faecalis V583 genome, there are two BSHs encoded by EF0521 and EF3005. In wild-type E. faecalis V583, the BSH encoded by EF0521 is induced at a higher level than EF3005, which suggests that the BSH encoded by EF3005 plays a minor role in the bile salt stress response. SlyA may in fact give a selective advantage to bacterial development in the intestine, as E. faecalis is naturally present in the gastrointestinal tract (Jones, Begley, Hill, Gahan, & Marchesi, 2008). However, other experiments using microarrays (Solheim, Aakra, Vebo, Snipen, & Nes, 2007) or a proteomic approach (Bøhle, et al., 2010) have produced contrary results, where the expression of these genes was not seen to be inducible by exposure to bile salt conditions. Differences in the results may be attributable to differences in experimental procedure or the detection limits of the various assays. The down-regulation of a number of genes was also observed, with a number of these being hypotheticals and with a majority related to protein synthesis (i.e. tRNA synthetases). However, expression of one of the tryptophanyl-tRNA- synthetases (EF2679, trpS) was found to be up-regulated at all time points studied, as compared to the initial start time. Further work is required to clarify the role of SlyA under different environmental conditions.
Regulation of E. faecalis Ebp pilus
The E. faecalis endocarditis and biofilm-associated pili (Ebp) is a major virulence factor that plays an important role in cell adhesion, biofilm formation, and the development of endocarditis. Many environmental factors affect the levels of Ebp surface expression, including culture media (TSB vs. BHI), serum, and bicarbonate levels, which suggests that this is a tightly regulated process (Nallapareddy, et al., 2006; Bourgogne, Thomson, & Murray, 2010). Three genes (ebpA, B and C) that encode the E. faecalis pilus structural proteins are located in the same polycistron, with the divergent promoter region shared with the ebpR gene.The ebpR gene encodes a protein that is a member of the AtxA/Mga family of transcriptional regulators. It has been demonstrated that the deletion of the ebpR gene leads to reduced levels of ebpABC mRNA and Ebp production, as evaluated by qRT-PCR and Western blot analysis, respectively (Bourgogne, Singh, Kox, Pflughoeft, Murray, & Garsin, 2007). As a result, the mutant ebpR gene displays phenotypes of reduced biofilm formation and attenuated primary adherence. In a recent study, Bourgogne et al. demonstrated that the addition of bicarbonate to culture media enhanced the expression of the ebpR and ebpABC loci. However, the authors did not observe a strong direct relationship between ebpR expression and ebpA expression, suggesting that the activation effect of bicarbonate to ebpABC may be through other unidentified factors other than ebpR (Bourgogne, Thomson, & Murray, 2010). Another recent study by Gao et al. identified RNase J2 as a positive regulator of Ebp expression (Gao, Pinkston, Nallapareddy, van Hoof, Murray, & Harey, 2010). Similar to ebpR, the deletion of E. faecalis rnjB resulted in a 30-fold decrease in ebpABC mRNA expression and significantly decreased Ebp surface display. RT-PCR and a reporter gene assay demonstrated that the mRNA level of ebpR is not affected by the deletion of RNase J2, excluding the possibility that RNase J2 affects ebpR at the RNA level through transcriptional regulation or RNA processing. It is likely, however, that RNase J2 affects epbR expression at the translational level through an as-yet unidentified regulatory or antisense RNA.
Intercellular Signaling Regulated Gene Expression
Quorum sensing is a global regulatory mechanism that bacteria use to communicate with each other through the accumulation and detection of signaling molecules that they release into the environment, which enables individual cells to respond to cell density (Ng & Bassler, 2009). Two major types of signaling molecules are used by different bacteria: Gram-negative bacteria produce N-acyl-homoserine lactones (HSLs) that bind to and activate a transcriptional activator, which in turn activates the transcription of regulated genes; while Gram-positive bacteria use extracellular peptides that are processed from bacteria-encoded proteins, through post-translational processes, as quorum sensing signaling molecules. Upon binding to the specific transmembrane receptors, these autoinducing peptides trigger signal transduction pathways, resulting in the activation or repression of effecter genes. In E. faecalis, several quorum-sensing–related regulation systems, which play important roles in biofilm formation, bacteria virulence, and production of cytolysin, toxin, have been identified (Haas, Shepard, & Gilmore, 2002; Hancock & Perego, 2004; Qin X. , Singh, Weinstock, & Murray, 2001). Two well-known examples, the Fsr and Cyl systems, are further discussed in detail.
One of the best-studied Gram-positive quorum-sensing systems is the staphylococcal Agr system. It consists of a transmembrane peptidase AgrB, the autoactivating peptide (AIP) processed from the propeptide AgrD by AgrB, and a classic two-component signaling module of AgrC (HPK) and AgrA (RR) (Novick & Geisinger, 2008). The homologous system in E. faecalis is the Fsr quorum sensing system. The E. faecalis fsr gene cluster consists of three genes, fsrA, fsrB, and fsrC, with a recent study revealing that the C-terminus of fsrB can be translated individually into a short peptide FsrD, which is the equivalent of AgrD (Nakayama, et al., 2006). Similar to the Agr system, FsrB is responsible for the processing and secretion of the signal peptide gelatinase biosynthesis-activating pheromone (GBAP) from FsrD. The extracellular accumulation of GBAP at high cell density allows binding to the membrane HPK FsrC, and triggers auto-phorsphorylation and dimerization of FsrC, which in turn transfers the phosphate group to the response regulator FsrA. The phosphorylated form of FsrA is able to bind to the promoter of the fsr gene cluster and activate the transcription of fsrBCD, resulting in the elevated production of GBAP for the further activation of fsr genes (Qin X. , Singh, Weinstock, & Murray, 2000). As demonstrated by Northern blot, the fsrC mRNA level in cultured E. faecalis is low during the early growth phase, but drastically increases in latter growth phases, confirming that fsr expression is dependent upon cell density.
Besides binding to its own promoter and self-activating through the positive feedback loop, the staphylococcal Agr system regulates transcription of a global antisense RNA (RNAIII), which in turn regulates expression of exoproteins and pleiotropic regulators at the translational level (Dunman, et al., 2001). Unlike the Agr system, the E. faecalis Fsr system does not have the RNAIII component as an effector, and apparently lacks the ability to broadly affect gene expression. In fact, some clinical and environmental E. faecalis strains carry a 23.9-kb deletion that covers much of the fsr gene locus (Nakayama, Kariyama, & Kumon, 2002). An interesting phenotype identified in some fsr mutant strains was that they were gelatinase-negative despite a gelE+ genotype. Qin et al. further demonstrated that deletion of the fsr gene cluster from the E. faecalis OG1RF significantly decreased the expression of two extracellular proteases, gelatinase (GelE) and serine protease (SprE), and that the regulation occurs at the transcriptions level (Qin X. , Singh, Weinstock, & Murray, 2000). As shown in Figure 2, the gelE/sprE genes are located immediately adjacent to the fsr gene cluster, though they are not cotranscribed with the fsr genes. A gelE-specific promoter (Pe) was detected by transcriptional fusion analysis, and has been identified as fsr-dependent (Qin X. , Singh, Weinstock, & Murray, 2001). Altogether, the Fsr system acts as an activator for the expression of gelE and sprE. Using a gene expression microarray approach, Bourgogne et al. further showed that the deletion of the fsrB gene affects the transcription of a number of other genes that encode EF1097, surface proteins EF0750-0757, and enzymes involved in several metabolic pathways (Bourgogne, Hilsenbeck, Dunny, & Murray, 2006). A recent report by Pinkston et al. reported a higher level of surface expression of the collagen adhesion protein, Ace, in fsrB mutants in the late growth phase, as compared to wild-type OG1RF (Pinkston, et al., 2011). They demonstrated that regulation of the Ace surface display by the Fsr system is mediated by gelatinase at the post-translational level: The activation of gelE expression at a high cell density by the Fsr quorum sensing induces gelatinase production, which in turn cleaves Ace from the cell-surface. Cleavage of Ace significantly inhibits the ability of the bacterial cells to adhere to collagen, which suggests a possible role of this regulation mechanism in E. faecalis colonization or dispersion.
Another example of a cell-density–dependent gene regulation mechanism comes from the autoinduction of cytolysin. Cytolysin is a peptide bacterial exotoxin expressed by some strains of E. faecalis. As a secreted virulence factor of E. faecalis, cytolysin is able to lyse eukaryotic cells, as well as other bacteria that lack the ability to produce cytolysin (Booth, Bogie, Salh, Siezen, Hatter, & Gilmore, 1996). The gene operon involved in the synthesis and regulation of E. faecalis cytolysin consists of eight genes: cylLL, cylLs, cylM, cylB, cylA, cylI, cylR1, and cylR2 (Haas, Shepard, & Gilmore, 2002). The cylLL and cylLs encode the large and small subunits of cytolysin, respectively, which are further secreted from the cell and processed by gene products of cylM, cylB, and cylA. The divergently transcribed cylR1 and cylR2, however, are not directly involved in the production and maturation of cytolysin peptides. Instead, they compose a novel two-component system that regulates gene expression of cytolysin in response to environmental cues, such as quorum signals or the presence of target cells. Although it has no apparent similarity to the classic two-component systems of HPK/RR pair, cylR1 functions as the membrane-associated sensor unit, while the DNA-binding protein cylR2 acts as a response regulator to alter the promoter activity of cytolysin-associated genes. This signal transduction machinery is responsible for the extracellular levels of one of the cytolysin peptides, cylLs. Expression of the cytolysin operon is repressed at subthreshold levels of the autoinducer cylLs (Coburn, Pillar, Jett, Haas, & Gilmore, 2004). When cylLs accumulates to threshold levels as a result of either higher cell density or the presence of a target cell which preferentially absorbs the other cytolysin peptide cylLL, derepression occurs leading to high-level cytolysin expression. This quorum-related regulation mechanism provides enterococci with a convenient tool to actively probe and respond to the environment.
Antimicrobial Response
E. faecalis V583 was the first vancomycin-resistant clinical isolate of E. faecalis reported in the United States, and was originally isolated from a patient suffering from a persistent bloodstream infection (Sahm, et al., 1989). V583 is rich in numerous virulence traits that have been acquired by horizontal gene transfer, which contribute to its success as a hospital pathogen (reviewed in (Paulsen, et al., 2003)). In addition to the acquisition of virulence genes which allow the enterococci to cause infection, V583 is one of the leading causes of hospital-associated Gram-positive infections, through its acquisition of genes that aid in maintaining antibiotic resistance. Antibiotics of various classes induce cell death by targeting different cellular processes, although we have a limited understanding of the extent and effect these antibiotics have on the cell besides binding to or inhibiting their known targets. The response of E. faecalis to various antibiotics most likely involves the coordinated interplay between the regulation of virulence factors and multiple genetic traits that govern adaptation of the bacterial cell’s physiology. Many transcriptomic studies of enterococci have focused on the response of V583 to different environmental cues. As enterococci continue to be a cause of hospital-acquired infections, it is particularly important to understand the effect of antibiotic pressure. Several studies have been conducted that examined the response of enterococci to various antimicrobial agents, including erythromycin, chloramphenicol, vancomycin, copper, and bacteriocins.
Erythromycin
Erythromycin is a macrolide antibiotic which acts by reversibly binding to the P site of the 50S subunit of the bacterial ribosome. Resistance mechanisms identified in Gram-positives to subvert the inhibitory effects of erythromycin include target modification by methylation, which changes the P site of the rRNA, as well as a macrolide efflux resistance mechanism (Pechère, 2001). E. faecalis V583 carries a plasmid known as pTEF1 that encodes an rRNA adenine dimethylase family protein (EF0007), which may aid in its resistance to this drug.
E. faecalis strain V583 is resistant to relatively high concentrations of erythromycin; however, the treatment does retard its growth. A drastic change in gene transcription was seen with erythromycin-treated V583, as compared to untreated cells, over a 90 minute period (Aakra Å. , et al., 2005). In the presence of 50 μg/ml of erythromycin (V583 is resistant to > 400 μg/ml), Aakra et al. (Aakra Å. , et al., 2005) found that 260 genes were down-regulated at one or more time points, while 340 genes were up-regulated. From this set, genes encoding for hypothetical proteins, as well as genes encoding for transport and binding proteins, were the two most dominant groups of differentially expressed genes. Surprisingly, in this experiment, the EF0007 gene that encodes for an rRNA adenine dimethylase family protein (ErmB), was not up-regulated in the presence of erythromycin, but was constitutively expressed in V583 even in the absence of erythromycin. This would suggest that the expression of genes other than ermB is important for the survival and growth of V583 in the presence of erythromycin.
Although changes in transcriptional levels of many genes with differential expression identified upon erythromycin exposure is assumed to occur mainly due to a change in growth conditions (Aakra Å. , et al., 2005), a number of these changes could be linked to survival in the presence of erythromycin. For example, the induction of ribosomal proteins and two protein Ala-acetyltransferases indicate that the overproduction of ribosomal proteins is one way to evade the effects of erythromycin. Interestingly, a MsrC-like protein (EF1413) was up-regulated at all time points studied. MsrC homologs found in E. faecium are thought to encode an efflux pump that is involved in low-level macrolide resistance (Portillo, Ruiz-Larrea, Zarazaga, Alonso, Martinez, & Torres, 2000; Singh, Malathum, & Murray, 2001). However, when compared to the MrsC homolog from E. faecium, the protein sequence of EF1413 from E. faecalis V583 is found to be only 40% identical (60% similar). Other specific genes found to be up-regulated were genes that encode ABC transporters that belong to the MDR family (EF1732 and EF1733) (Aakra Å. , et al., 2005). Another MDR efflux pump, EmeA (EF1078), is also induced at all time points, excluding time zero in V583 exposed to erythromycin, and is most likely involved in mediating resistance. This finding supports previous observations that the efflux pump could extrude erythromycin (Jonas, Murray, & Weinstock, 2001). A number of two-component signal transduction pathways were also found to be up- or down-regulated in the presence of erythromycin—although further work is required to clarify their roles in E. faecalis. It is assumed that these factors all play some role in mediating erythromycin resistance.
Another interesting observation made by Aakra et al. (Aakra Å. , et al., 2005) is that in the presence of erythromycin, six genes related to fatty acid and phospholipid metabolism were up-regulated. In Gram-negative bacteria, such intrinsic resistance to macrolides is achieved due to the impermeability of the cellular outer membrane to this hydrophobic drug (Roberts M. C., 2008), and it may be that changes in membrane biochemistry in enterococci also impede the entry of this class of drug into the cell.
Chloramphenicol
Chloramphenicol is a broad-spectrum antibiotic that inhibits the peptidyl transferase reaction of the large subunit of the ribosome. This antibiotic currently has limited use in treating bacterial infections in humans, due to widespread acquisition of resistance mechanisms. Bacterial resistance to chloramphenicol may be mediated by several mechanisms, including inactivation by an enzyme (acetyltransferase or phosphotransferase), the excretion of the drug by an efflux pump, and mutations in the 23S rRNA that result in an altered target site in the cell. The well-studied E. faecalis strain V583 is resistant to chloramphenicol, but no specific chloramphenicol resistance genes have been identified in the genome sequence, and resistance cannot be solely attributed to mutations in the 23S rRNA gene. Interestingly, the growth of V583 in the presence of chloramphenicol is decreased, which suggests that chloramphenicol does create stress on the cells. In the presence of chloramphenicol, over 609 genes were up-regulated and down-regulated at one or more of three time points (t=0, 90, and 180 minutes), and these genes were identified through two different methods to undergo transcriptional changes (Aakra A. , et al., 2010).
In response to the addition of chloramphenicol to the growth medium, it would be expected that the bacterial cells would want to adapt to the conditions, potentially through changes in the cell wall, as was observed for treatment with other agents, such as sodium dodecyl sulfate (Solheim, Aakra, Vebo, Snipen, & Nes, 2007) and erythromycin (Aakra Å. , et al., 2005). Two genes (ef1732 and ef1733) that encode potential transporters of chloramphenicol out of the V583 cells were strongly induced in cells treated with the drug at 90 minutes. These genes encode for an ABC transporter that belongs to the MDR family of transporters, and were also seen to be up-regulated in the presence of erythromycin (Aakra Å. , et al., 2005). Genes involved in fatty acid biosynthesis were up-regulated in E. faecalis V583 cells at 180 minutes (Aakra A. , et al., 2010), and a similar response was noted for the growth of E. faecalis in the presence of sodium dodecyl sulfate (Solheim, Aakra, Vebo, Snipen, & Nes, 2007) and erythromycin (Aakra Å. , et al., 2005). The up-regulation of these genes suggests that V583 is adapting to the stress induced by chloramphenicol by altering the membrane, which may change the transport capabilities of the cell as a result. This change in membrane composition may also partly explain the relatively low number of induced genes that specify transport and binding proteins. Interestingly, a number of phage genes were also up-regulated in the cells exposed to chloramphenicol, potentially indicating that the potential for cell lysis induces the phage to leave the cell. There are a number of phage-encoded genes in the V583 genome, including one phage that is found in many different E. faecalis strains (phage 02); however, the transcriptional profile for the phage 02 genes was not found to change in the time course of chloramphenicol exposure.
The genome of V583 carries the vanB operon, which encodes for resistance to vancomycin. In the presence of chloramphenicol, 8 of the 11 genes encoded in the operon were induced, including the genes encoding for the histidine kinase and the cognate response regulator, which are responsible for sensing vancomycin and inducing the expression of the rest of the operon.
The addition of chloramphenicol to the medium led to the up-regulation of the pyrimidine biosynthesis operon (ef1721 to ef1712). Interestingly, the precursor for pyrimidine synthesis, carbamoyl phosphate, is also the precursor of arginine biosynthesis. Arginine biosynthesis in chloramphenicol-treated V583 cells was down-regulated (Aakra A. , et al., 2010). This response is different than what was seen in erythromycin exposed E. faecalis V583 (Aakra Å. , et al., 2005), suggesting that it is specific to the presence of chloramphenicol.
As the target of chloramphenicol is the ribosomal protein, it was hypothesized that transcriptional changes would be seen in a number of ribosomal proteins. Although the increased transcription of the ribosomal proteins were noted by Aakra et al. (Aakra A. , et al., 2010), it is also important to keep in mind that microarray analysis may not actually reflect the level of ribosomal proteins in the cells, as many proteins may also be regulated at the translational level.
Vancomycin
Sub-inhibitory concentrations of vancomycin are known to trigger changes in cell wall biosynthesis genes in Gram-positive bacteria, including in E. faecalis. Vancomycin is known to inhibit cell wall synthesis by covalently binding to the D-alanyl-D-alanine chain of the cell wall pentapeptide (Reynolds P. E., 1989). Vancomycin resistance in E. faecalis and E. faecium can be mediated by either the vanA or vanB operon. Both operons encode for the expression of polypeptides that are involved in aiding in the transposition of the transposon that encodes the van genes, as well as genes encoding for the modification of the cell wall pentapeptide to prevent vancomycin from binding to its target (reviewed in (Courvalin, 2006)). The difference between the vanA and vanB operons is that the vanA operon can also be induced by the related glycopeptide antibiotic teicoplanin (Baptista, Depardieu, Reynolds, Courvalin, & Arthur, 1997; Baptista, Rodrigues, Depardieu, Courvalin, & Arthur, 1999).
Transcription of the vancomycin-resistant gene cluster is regulated by the VanRS two-component system that is found at the beginning of the gene cluster (Arthur, Molinas, & Courvalin, 1992; Evers & Courvalin, 1996). Phosphorylation of the VanR response regulator enhances the affinity of the protein for the regulatory regions of the promoter regions of the vanA gene cluster; namely, for vanRS (PR) and vanHAXYZ (PH) (Arthur, Depardieu, Gerbaud, Galimand, Leclercq, & Courvalin, 1997; Holman, Wu, Wanner, & Walsh, 1994). A similar setup is also found to regulate the expression of the vanB gene cluster (Baptista, Depardieu, Reynolds, Courvalin, & Arthur, 1997; Arthur, Depardieu, Gerbaud, Galimand, Leclercq, & Courvalin, 1997; Wright, Holman, & Walsh, 1993). The response regulators of both van operons can also be activated by kinases encoded by the host chromosome in the absence of their partner sensor (Baptista, Rodrigues, Depardieu, Courvalin, & Arthur, 1999; Arthur, Depardieu, Gerbaud, Galimand, Leclercq, & Courvalin, 1997; Silva, Haldimann, Prahalad, Walsh, & Wanner, 1998).
The activation of the promoters in the vanA operon by VanR is inducible by vancomycin, teicoplanin, and moenomycin in wild-type strains of E. faecalis, and these are constitutively expressed in vanS mutants (Arthur, Depardieu, Gerbaud, Galimand, Leclercq, & Courvalin, 1997). The vanB cluster differs with the operon being inducible only by the addition of vancomycin, whereas in a vanSB mutant, vancomycin, teicoplanin, and moenomycin can all activate transcription of the operon from the two promoter sites (Baptista, Rodrigues, Depardieu, Courvalin, & Arthur, 1999; Evers & Courvalin, 1996). Activation of the vanA operon by glycopeptides and moenomycin occurs as these drugs inhibit the transglycosylation reaction, but by different mechanisms, as the structure of moenomycin is unrelated to that of the glycopeptides (Handwerger & Kolokathis, 1990). It is thought that the VanSA sensor kinase in the vanA operon is not involved in direct interaction with the stimulating drugs (Handwerger & Kolokathis, 1990). With the vanB operon, the VanSB sensor kinase does appear to interact with vancomycin, which distinguishes the transcriptional differences between these two operons. Amino acid substitution of the N-terminal sensing domain of VanSB allows for the induction of the vanB operon by the other glycopeptide teicoplanin, but not by moenomycin (Baptista, Depardieu, Reynolds, Courvalin, & Arthur, 1997; Baptista, Rodrigues, Depardieu, Courvalin, & Arthur, 1999). Experiments completed both in vitro and in vivo have shown that carriage of the transposon encoding for the vanB operon, when uninduced, was not significantly costly to the host (Foucault, Depardieu, Courvalin, & Grillot-Courvalin, 2010). Induction of the resistance genes leads to a major reduction in growth rate, colonization, and transmission in Enterococcus sp. (Foucault, Depardieu, Courvalin, & Grillot-Courvalin, 2010). Overall, tight regulation mediated by a two-component regulatory system effectively reduces the biological costs that are associated with vancomycin resistance.
Experiments that used sub-inhibitory concentrations of vancomycin on the E. faecalis strain V583, which carries the vanB operon, revealed that the open reading frame EF2292 (named vanV) located 3’ to the vanX gene exhibited the same level of induction as the vancomycin resistance genes (Ribeiro, Santos, Marques, Gilmore, & de Fátima Silva Lopes, 2011). Analysis of this gene suggests that it might be involved in some regulation process of the vanB operon in strains that carry vanV, although it is not consistently found in all vanB-carrying strains. It also appears that vanV does not have an effect on the resistance of the carrying strain to vancomycin (Ribeiro, Santos, Marques, Gilmore, & de Fátima Silva Lopes, 2011). Further screening of vanB-resistant enterococci for the presence of this gene is required, and may unravel a particular function for this gene.
It should be noted that many enterococcal species can carry other van operons, which encode for low-level or constitutive resistance to vancomycin. One of these operons is the vanD operon, which encodes for the constitutive production of the peptidoglycan precursor ending in D-alanyl-D-lactate (Depardieu, Bonora, Reynolds, & Courvalin, 2003). Mutations in the sensor kinase and response regulator, as well as the native D-alanyl-D-alanine ligase (involved in native cell wall synthesis), lead to the constitutive expression of the van operon and the cell wall that contains only the D-alanyl-D-lactate pentapeptide structure (Depardieu, Reynolds, & Courvalin, 2003; Depardieu, Kolbert, Pruul, Bell, & Courvalin, 2004).
Another van operon carried by E. casseliflavus, E. gallinarum, and E. flavescens is the vanC operon. The vanC phenotype is inducible and constitutive, and results in the production of a peptidoglycan precursor ending in D-Alanyl-D-Serine (Reynolds & Courvalin, 2005). Other van operons that encode for a pentapeptide ending in a serine residue are found in E. faecalis and are referred to as the vanE and vanG operons. The vanE operon has an organization identical to the vanC operon (Patiño, Courvalin, & Perichon, 2002). The vanG operon has a unique gene organization and appears to encode a predicted transcriptional activator upstream of the two component regulatory system (Depardieu, Bonora, Reynolds, & Courvalin, 2003).
The expansion of the van operon alphabet is continually expanding with vanL (Boyd, Willey, Fawcett, Gillani, & Mulvey, 2008), vanM (Xu, et al., 2010), and vanN (Lebreton, et al., 2011) gene clusters being reported. Transcriptional regulation of these operons appears to be similarly regulated by a two component transduction system and promoters, as in the van operons discussed above.
Bacteriocins
Bacteriocins are peptides secreted by bacteria with the intent of inhibiting the growth of similar or closely related bacterial species. Many lactic acid bacteria produce bacteriocins that have the ability to inhibit the bacterium beyond the genus level, and also have the potential to be used medically for the prevention of growth of unwanted microflora. With the spread of antibiotic resistance, bacteriocins are ideal candidates for use as alternative antimicrobials. In addition, resistance to bacteriocins is not common in nature. Class IIa bacteriocins encompass the non-lantibiotic bacteriocins that have regular amino acid residues. A well-characterized class IIa bacteriocin is pediocin, which is produced by Pediococcus acidilactici (Marugg, et al., 1992). Many of these bacteriocins are produced by lactic acid bacteria and have been found to inhibit other Gram-positive organisms, including Enterococcus spp. Bacteriocins act by permeabilizing the cell membrane, which leads to the killing of the cell. The target for class IIa bacteriocins is the mannose phosphate transport (mpt-PTS), and mutants that lack this system are insensitive to the effects (Diep, Skaugen, Salehian, Holo, & Nes, 2007; Gravesen, et al., 2002; Héchard & Sahl, 2002; Ramnath, Beukes, Tamura, & Hastings, 2000). The PTS is a major uptake system for mannose and glucose, and many of the components are also involved in gene regulation of catabolic operons. In the laboratory, deletion mutants of mptR and the mpt operon are known to be resistant to the effects of bacteriocins.
Transcriptional analysis has been completed on pediocin-resistant strains of E. faecalis generated in the laboratory environment to test the efficacy of this bacteriocin in the treatment of hospital-acquired enterococcal infections. A comparison of wild-type V583, spontaneously resistant isolates, and an ΔmptD mutant, which is known to render E. faecalis strains resistant to the effects of the class IIa bacteriocin pediocin, was completed (Opsata, Nes, & Holo, 2010). In E. faecalis, the mpt operon is under transcriptional control of a promoter recognized by σ54, and is dependent upon the activator MptR. The spontaneous bacteriocin resistant isolates contained a mutation in mptR (EF0018; A356G mutation) causing the down-regulation of the expression of the mpt operon. The expression of several other genes were also down-regulated, and included the components of the mpt operon (EF0019-EF0022); the downstream mannose operon regulator (manO; EF0024); a major facilitator family transporter (EF0082); glyceraldehyde-3-phosphate dehydrogenase (gap-2; EF1964); phosphoglycerate kinase (pgk EF1963); triosephosphate isomerase (triA; EF1962); enolase (eno, EF1961); phosphoglycerate mutase (gpm,EF0195); pyruvate kinase (pyk; EF1046); and L-lactate dehydrogenase (ldh-1; EF0255); a majority of which encode for sugar metabolism. A study by Opsata et al. (Opsata, Nes, & Holo, 2010) found the reduced consumption of glucose as a carbon source in the mpt mutants, which demonstrated the important role of Mpt in glucose metabolism in E. faecalis. The reduced glucose uptake results in changes in the concentrations of glycolytic metabolites, which subsequently affects the energy status of the cell. Putative carbon catabolite regulation (cre)-sites were located upstream of three of the differentially expressed genes, and some with cre-sites were located downstream of the genes, supporting the idea that an altered energy status sensed by a histidine protein-kinase/phosphorylase and implemented on the PTS phospho-carrier protein does occur. However, this is also the first time that cre-sites have been reported downstream of the regulated gene (Opsata, Nes, & Holo, 2010). In E. faecalis, disruption of the mannose PTS altered the transcription of about 90 cre-sites, with about 65% of these being involved in the uptake and metabolism of alternative energy sources. A number of these genes were also found to encode transcriptional regulators, and may indicate that a regulatory cascade has been triggered. A number of the genes found to be differentially regulated in the E. faecalis V583 mpt bacteriocin-resistant mutants were also found to be similarly expressed in a transcriptome study of a Streptococcus mutans EIIAB mannose-PTS mutant; however, some differences were found that may be attributed to niche preferences (Opsata, Nes, & Holo, 2010).
Copper
Metal ions are crucial for bacteria to maintain transcriptional control of regulatory networks that govern gene expression. Maintaining homeostasis between the intra- and extracellular concentration of metal ions is critical in adaptations to intracellular survival and replication among pathogenic bacteria. Copper is useful as a cofactor for redox enzymes in reactions, due to its ability to accept and donate electrons; however, this very reactivity also makes it a toxic metal (Urbański & Beresewicz, 2000; Linder & Hazegh-Azam, 1996). It is therefore crucial for cells to maintain control over copper by means of uptake, storage, and efflux.
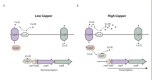
Copper uptake and regulation in E. hirae is a well-studied system. The regulation of intracellular copper in this species is mediated by the cop operon, which consists of four open reading frames (Solioz & Stoyanov, 2003) (Figure 3). The four genes encode two P-type ATPase copper transporters (copA and copB), a copper chaperone protein (copZ), and a copper-dependent transcriptional regulator (copY) (70). This operon is conserved between all members of the Lactobacillales and includes at least homologues of the copA and copY genes (Reyes, Leiva, Cambiazo, Méndez, & González, 2006).
The transporters in the cop operon are responsible for opposite processes. CopA takes up copper (I) when it is limiting, and structural information for this ATPase is available (Lübben, Portmann, Kock, Stoll, Young, & Solioz, 2009; Tsuda & Toyoshima, 2009). CopB extrudes copper (I) when it reaches toxic concentrations (Odermatt & Solioz, 1995; Solioz & Odermatt, 1995). CopY is a bipartite repressor, where the N-terminal half of the protein exhibits 20% identity to bacterial repressors of β-lactamases, such as MecI (Solioz & Stoyanov, 2003). The N-terminal region also contains a QQ motif found in the phage repressors (lambda and 434), and has been shown to tightly interact with an ACA triplet of the DNA binding site (Anderson, Ptashne, & Harrison, 1987). Mutations of the DNA binding site of CopY abolish the ability of the repressor to interact with DNA in vitro (Wunderli-Ye & Solioz, 1999). The C-terminal end of CopY has multiple cysteine residues arranged in a pattern commonly seen in other copper-responsive transcriptional activators that have been studied in yeast (CxCx3CxC) (Zhou & Thiele, 1991; Dobi, Dameron, Hu, Hamer, & Winge, 1995). CopY functions as a dimer (Strausak & Solioz, 1997), binding to an inverted repeat sequence upstream of the cop operon when bound to zinc (Zn (II)) (Cobine, Jones, & Dameron, 2002), suppressing transcription. Figure 3 illustrates the role of CopY in the presence of high copper concentrations and low (physiological) copper concentrations. CopZ or another appropriate copper complex can donate two copper (Cu(I)) to CopY, displacing the bound Zn(II), which results in CopY releasing itself from the DNA and allows expression of the downstream cop genes (Cobine, Jones, & Dameron, 2002). The interaction of CopY and CopZ carrying Cu(I) is dependent on electrostatic interactions, and transfer of the Cu(I) occurs due to affinity differences for this metal, as well as charged-based interactions between the two proteins (reviewed in (Solioz & Stoyanov, 2003)).An interesting detail pertaining to CopZ is that it more easily degraded when bound to Cu(I) by a cellular protease under high copper stress (Lu & Solioz, 2001). It is hypothesized that CopZ degradation prevents cell damage from occurring as a result of the exposed Cu(I) in the protein metal ion complex, which can participate in Fenton-type reactions leading to formation of reactive hydroxyl radicals and cell damage (Lu, Dameron, & Solioz, 2003; Solioz M. , 2002).
With the emergence of microarray technology, the effect of copper on the transcriptome of enterococci species has been more closely studied. Two recent publications have looked at the transcriptional response of E. faecalis OG1RF (Reyes-Jara, et al., 2010) and V583 (Abrantes, Lopes, & Kok, 2011) in the presence of copper. Not surprisingly in both studies, the cop operon (EF0297-EF0299) was the most up-regulated operon. In V583, in the presence of 0.05 mM CuSO4, other induced operons included the following: genes involved in potassium transport (kpdA and kpdB); an operon encoding for two ABC transporters, a hypothetical protein and a GntR family transcriptional regulator (EF1673-EF1676); an operon of V-type ATP enzymes (EF1492-EF1500); nine genes related to cell wall biosynthesis; and several virulence-associated genes, including LemA (EF0468), a cell-envelope associated acid phosphatase (EF3245), and a cell wall surface anchor family protein (EF3314) (Abrantes, Lopes, & Kok, 2011). In the study conducted by Reyes-Jara et al. (Reyes-Jara, et al., 2010), the authors found that an OG1RF ΔcopY mutants’ response to the presence of 0.5 mM CuSO4 was similar to the wild-type copY strain, with the exception of the cop operon being constitutively expressed in the absence of copper. Several other transcriptional elongation factors were also induced shortly after the CuSO4 exposure, including GreA (EF2914), Rrf2 (EF3175), the Cro/CI (EF0873), Sor/DeoR (EF1965) and a hypothetical protein of COG class K (EF1752). The induction of these genes may not occur as a result of increased copper concentration, but may be induced in response to a secondary signal, such as the oxidative stress generated by copper exposure. Comparison between the two strains’ responses to copper is difficult, given that the experimental conditions were different.
Response to Environmental Cues
The bacterial transcriptome is a dynamic entity that closely reflects the organism's response to the environmental conditions in which it resides. DNA microarray-based transcriptional profiling provides a genome-wide portrait of the transcriptome of the organism, and may disclose important clues on how a bacterium adapts to a particular environmental niche as a result. Much of the effort put into enterococcal transcriptome analysis has consequently focused on stressors relevant to the human host. In vitro experimental approaches using a range of concentrations of various body fluids have been carefully designed in an attempt to mimic in vivo scenarios, and these studies may provide essential information about the mechanisms involved in an important aspect of the enterococcal life cycle. Indeed, Snyder et al. (Snyder, et al., 2004) observed a considerable overlap between the expression profiles obtained from Escherichia coli during colonization of the murine urinary tract and during growth in human urine. In the following section, we will discuss the transcriptional responses of E. faecalis (mainly the sequenced V583 strain) when exposed to relevant body fluids.
Bile
Bile is a digestive juice secreted by the liver and stored in the gallbladder, and it functions both to assist the digestion and absorption of fat in the gut and as a means for the body to excrete waste from the blood. The concentrations of bile present in the small intestines range from 0.2–2%, but can get up to 8% in the gall bladder (Bowen, 1998). The ability to resist the deleterious action of various bile components is thus an absolute need for E. faecalis in its commensal lifestyle. The mechanisms involved in bile resistance in E. faecalis V583 were previously assessed in a genome-wide transcriptional analysis over a time course of 120 min. after the addition of 1% bovine bile to the growth medium (Solheim, Aakra, Vebo, Snipen, & Nes, 2007). The detergent-like action of bile acids was reflected as an enrichment of V583 genes that code for proteins with membrane-associated functions and/or locations among the differentially transcribed genes. In particular, the functional categories of genes involved in fatty acid and lipid metabolism and signal transduction were strongly affected. Studies in lactobacilli (Bron, Molenaar, de Vos, & Kleerebezem, 2006; Pfeiler, Azcarate-Peril, & Klaenhammer, 2007; Whitehead, Versalovic, Roos, & Britton, 2008) and Bifidobacterium breve (Ruiz, Zomer, O'Connell-Motherway, van Sinderen, & Margolles, 2012) also suggest that cell envelope stress is one of several aspects of the bile response that is conserved among Gram-positive bacteria.
Active extrusion of bile appears to be another common strategy to counteract bile toxicity in bacteria (Storz, 2000). Transcriptional profiling has previously been used to identify bile efflux proteins in Lactobacillus reuteri and B. breve (Whitehead, Versalovic, Roos, & Britton, 2008; Ruiz, Zomer, O'Connell-Motherway, van Sinderen, & Margolles, 2012), for example. Several candidate transporters were also reported in E. faecalis V583, including two members of the EmrB/QacA subfamily of the major facilitator superfamily of multidrug resistance transporters (Solheim, Aakra, Vebo, Snipen, & Nes, 2007). In E. coli, EmrB was shown to play a role in bile resistance and efflux of bile (Lomovskaya & Lewis, 1992; Thanassi, Cheng, & Nikaido, 1997), but the study of Solheim et al. (Solheim, Aakra, Vebo, Snipen, & Nes, 2007) was the first report of a proton motive force dependent transport system involved in bile resistance in Gram-positive bacteria. Shortly after this study, however, a similar induction of a gene coding for an EmrB/QacA-like multidrug resistance transporter (lr1584) was observed in L. reuteri (Whitehead, Versalovic, Roos, & Britton, 2008). Interestingly, lr1584 is part of an operon structure which was up-regulated during bile exposure in Lactobacillus acidophilus, as well as in L. reuteri and E. faecalis. Inactivation of both lr1584 and lr1582 resulted in an impaired adaptation phenotype in L. reuteri. Another V583-encoded transport system (EF2641-42) with significant homology to the well characterized bile responsive bilE system in Listeria monocytogenes (Sleator, Wemekamp-Kamphuis, Gahan, Abee, & Hill, 2005), also displayed differential expression in V583. This operon codes for proteins involved in glycine betaine uptake. The role of compatible solutes in bacterial stress management, such as glycine betaine, was recently reviewed by Sleator and Hill (Sleator & Hill, 2010).
Bile salt hydrolases (BSH) have also been implicated in bile resistance in other Gram- positive bacteria. These enzymes have been proposed to confer the ability to detoxify bile salts, and studies with a Δbsh mutant revealed an important role for BSH in the intestinal persistence of L. monocytogenes (Begley, Sleator, Gahan, & Hill, 2005). None of the two bsh homologs in V583 (EF0521 and EF3005) showed significant differential expression during exposure to bovine bile (Solheim, Aakra, Vebo, Snipen, & Nes, 2007); however, the bile-sensitive phenotype of an E. faecalis ΔslyA mutant was recently connected to the regulation of ef3005 by the transcriptional regulator SlyA (Michaux, Martini, Hanin, Auffray, Hartke, & Giard, 2011), which suggests the potential role of BSH in E. faecalis bile response after all. The diverging result can most likely be attributed to the increased sensitivity associated with real-time PCR, as compared to the microarray-based approach. As a result, further characterization of the loci in question will be needed to conclude whether BSH constitutes a mechanism for bile resistance in E. faecalis.
Urine
The urinary tract is among the most common sites of bacterial infection in humans, and urinary tract infections (UTIs) are the type of infection most commonly caused by enterococci (Malani, Kauffman, & Zervis, 2002). In order to identify genetic traits that may distinguish pathogenic and non-pathogenic E. faecalis in their ability to cause UTIs, the global transcription signatures of a probiotic (Symbioflor 1) and two pathogenic (MMH594 and OG1RF) strains during cultivation in human urine were compared (Vebø, Solheim, Snipen, Nes, & Brede, 2010). The non-pathogenic strain grew at the same rate as the pathogenic strains, which indicates that the pathogenic potential is not related to newly required mechanisms that enable growth in this environment. Although the three strains have different strategies in relation to their host, their transcriptional signatures towards urine as a growth medium were surprisingly similar. However, some potentially significant differences were reported. The highest number of differentially expressed genes was observed after 5min in OG1RF, whereas the process took 30min in MMH594 and Symbioflor 1. Consequently, it may be inferred that OG1RF adapts more rapidly to growth in urine. This rationale was further supported by the swift derepression of genes involved in macromolecular biosynthesis (i.e. transcription and protein synthesis) in OG1RF, as compared to the two other strains.
More than 50% of all microbial infections have been associated with the formation of biofilms, and biofilm formation appears to also be clinically relevant for UTIs. A particularly problematic aspect of biofilm formation is the ability of sessile bacteria to withstand host defense mechanisms and the increased resistance to antibiotics, biocides, and hydrodynamic shear forces observed for bacteria in biofilm (Bitsori, Maraki, Raissaki, Bakantaki, & Galanakis, 2005). Several genetic determinants with impact on biofilm formation in E. faecalis were differentially transcribed during growth in human urine, as compared to the rich medium 2×YT: bopABCD (biofilm on plastic; EF0954 to -57) this process was partially up-regulated in OG1RF, while srtA (EF3056) was induced in MMH594 and Symbioflor 1. Data from knock-out mutant studies also inferred a role for salA (EF3060), salB (EF0394) and altA (EF0799) in biofilm production, all of which were down-regulated in response to urine (Vebø, Solheim, Snipen, Nes, & Brede, 2010; Mohamed, Tenq, Nallapareddy, & Murray, 2006).
SalB was initially characterized as a stress-inducible protein in E. faecalis (Breton, Mazé, Harte, Lemarinier, Auffray, & Rincé, 2002). Its growth in urine also affected the expression of a large number of other genes with a proven or predicted function in stress responses in E. faecalis. One of the most pronounced effects was the stimulation of an oxidative stress response. Mn2+-depletion has been reported as a regulator of oxidative stress regulons in other bacteria. Interestingly, the pathogenic strain MMH594 seemed slightly better equipped with genes involved in manganese acquisition than the two other isolates: while efaABC and two other genes encoding Mn2+/Fe2+ transporters (EF1057 and EF1901) were induced during growth in human urine in all three strains, the putative uptake system for manganese encoded by EF0575 to -78, located on the pathogenicity island, is specific to MMH594. Vebø and coworkers reported the latter system to be induced in MMH594 during cultivation in urine (Vebø, Solheim, Snipen, Nes, & Brede, 2010). Taken together with the differential regulation of several iron transporters, these observations may be indicative of urine as an iron- and manganese depleted environment. This hypothesis was further supported by up-regulation of genes involved in the uptake and assimilation of iron in E. coli during growth in the urinary tract (Snyder, et al., 2004).
Surface structures are major determinants of virulence for many pathogens: namely, the presence of a capsule may allow the microorganism to evade the host immune system. Indeed, several cell wall polysaccharides with implications in the pathogenesis of enterococcal infections have been reported (Hancock & Gilmore, 2002; Huebner, et al., 1999; Xu, Murray, & Weinstock, 1998; Xu, Singh, Murray, & Weinstock, 2000), two of which showed differential expression upon the encounter with urine. Both the cps (capsular polysaccharide) and the epa (enterococcal polysaccharide) loci were down-regulated during growth in urine. The data from the transcriptional analysis also suggested that E. faecalis adjusts the fatty acid and phospholipid composition of the membrane in response to urine as a growth medium. This was manifested as an up-regulation of two gene clusters (EF0282-84 and EF2886-75) that were responsible for type II fatty acid biosynthesis (FASII) and isomerization of membrane phospholipids (Vebø, Solheim, Snipen, Nes, & Brede, 2010).
Urine is a complex medium consisting of a mixture of carbon sources (Tasevska, Runswick, McTaggart, & Bingham, 2005; Wishart, et al., 2009) that are likely to change over time, and the ability of the bacteria to exploit available carbon and energy sources may be crucial for virulence. The final cell density reached by E. faecalis in urine compared to those in 2× YT growth medium suggests that important growth factors are absent in urine; however, supplementary growth experiments disproved glucose as a major limiting factor for growth of E. faecalis in urine, as elevated glucose levels alone did not significantly augment bacterial growth (Solheim, unpublished). Nevertheless, the induction of EF3215 to -22 pointed towards citrate as a significant carbon source for E. faecalis during growth in urine. The cit operon is responsive to carbon catabolite repression (CCR) (Blancato, Repizo, Suárez, & Magni, 2008; Suárez, Blancato, Poncet, Deutscher, & Magni, 2011), and this observation is indicative of low glucose levels in urine. Furthermore, the transcriptional data suggested that E. faecalis uses available peptides as a source of amino acids.
The encounter with urine also had a considerable impact on the expression of a number of genes with a potential implication in virulence; however, among the established enterococcal virulence traits, only the fsr operon displayed significant changes in transcription. From the microarray data, a modest up-regulation of the fsrABC genes (EF1822 to -20) was seen in MMH594 at t30. The fsrA gene was also up-regulated at t5. The regulation of fsrB (EF1821) in MMH594 at t30 was, however, not confirmed by real time quantitative PCR (QPCR)—thus the importance of this observation remains uncertain. On the other hand, QPCR revealed significant up-regulation of fsrB in OG1RF after 30 minutes exposure to urine. As mentioned previously, the fsr system modulates expression of gelE-sprE operon through a classic quorum sensing signaling cascade. Secretion of these proteases that damage host tissue may contribute to bacterial migration and invasion. The gelE gene has been shown to be predominant among clinical enterococcal isolates; however, contradictory results have later been reported (Roberts, Singh, Okhuysen, & Murray, 2004; Coque, Patterson, Steckelberg, & Murray, 1995). Moreover, isogenic strains of E. faecalis divergent in gelatinase and serine protease production exhibited attenuated killing in nematode- and mice infection models (Qin X. , Singh, Weinstock, & Murray, 2000; Sifri, et al., 2002). An up-regulation of the gelE-sprE operon after prolonged exposure to urine is in line with the work by Shepard and Gilmore (115) which used QPCR to show that gelE transcription was 7-fold induced during logarithmic growth in urine.
Blood
E. faecalis is also a significant cause of hospital-acquired bacteremia (Malani, Kauffman, & Zervis, 2002; Wisplinghoff, Bischoff, Tallent, Seifert, Wenzel, & Edmond, 2004), and with this in mind, Vebø et al. conducted a whole-genome transcription study on the adaptation of E. faecalis to blood (Vebø, Snipen, Nes, & Brede, 2009). Interestingly, several aspects of the transcriptional signature of E. faecalis grown in blood were similar to that of E. faecalis grown in urine. An activation of many of the same genes implicated in stress responses was observed, with an overrepresentation of genes involved in oxidative stress adaptation (e.g. ohr, npr, sodA, ahpFC and nox). Moreover, many of the observed changes in gene expression could also be related to an extensive remodeling of the cell envelope. The changes detected among genes involved in FASII were also particularly pronounced. Other differentially transcribed operons with membrane-associated functions included lrgAB (EF3194-93), cps (EF2492-84), epa (EF2200-2189 and EF2184-77), and dltABCD (EF2749-46). D-alanylation of cell wall teichoic acids and lipoteichoic acids by the Dlt complex leads to a reduced negative charge of the bacterial surface. In this case, the reduced content of D-alanine esters in the teichoic acid may have resulted in an increased net negative charge on the bacterial cell surface, which in turn may have affected several bacterial properties, such as susceptibility to cationic antimicrobial peptides and biofilm formation (Theilacker, et al., 2006). The microarray data also showed that genes related to iron acquisition represented one of the major changes related to E. faecalis’s adaptation to blood, despite the fact that iron is not essential for the growth of most lactic acid bacteria. A similar trend was also observed during growth in urine, but the transcriptional changes provoked by blood were more pronounced. The induction of the genes responsible for citrate metabolism was observed in both the blood and urine experiments. In addition, genes involved in the breakdown of other CCR regulated substrates also displayed enhanced transcription in blood, including arginine (Barcelona-Andrés, Marina, & Rubio, 2002) and glycerol (Opsata, Nes, & Holo, 2010), along with possibly other C3-glycerides present in blood. As a result, the glucose concentration in body fluids such as blood and urine appears to be below the threshold for release of CCR. As for the established enterococcal virulence factors, QPCR analysis showed that fsrB and gelE were down-regulated in response to growth in blood, the opposite of what was observed in urine. Whereas cues present in urine stimulate fsr expression, factors present in blood seemingly interfere with its expression. This is the opposite effect from growth in serum by the E. faecalis MMH594 strain (Shepard & Gilmore, 2002).
Ethanolamine—An Interesting Case of Post-Transcriptional Regulation
E. faecalis is able to utilize ethanolamine as a source of both carbon and nitrogen, as recently reviewed (Garsin, 2010). This nutrient is abundantly present in the intestinal tract from sloughed-off epithelial cells, resident microflora, and ingested, processed foods. The E. faecalis ethanolamine utilization (eut) genes are encoded in a complex locus of 18 genes and include structural, enzymatic, and regulatory components. Upregulation of eut genes is triggered by the presence of two environmental cues—ethanolamine and adenosylcobalamin (AdoCbl). Each of these input signals is perceived and relayed by two different regulatory systems, and synchrony between them is required for expression of the eut genes. Both regulatory systems, as described below, control gene expression at the post-transcription initiation level and target nascent RNA instead of DNA. One is a cis-acting RNA structure, while the other is a response regulator (Garsin, 2010).
EutV, an ANTAR-family member with RNA-binding antiterminator activity
Upregulation of the E. faecalis eut genes is partially controlled by a two-component system. This consists of a soluble histidine kinase, eutW, and its cognate response regulator, eutV. Experiments showed that this two-component system specifically senses ethanolamine. In vitro, purified eutW underwent autophosphorylation in the presence of ethanolamine and was also able to transfer the phosphate residue to eutV (Del Papa & Perego, 2008; Fox, et al., 2009). In vivo, eut gene expression was demonstrated to be dependent on ethanolamine, eutV, and eutW (Del Papa & Perego, 2008; Fox, et al., 2009; Baker & Perego, 2011; Ramesh, et al., 2012). EutV belongs to the AmiR and NasR transcriptional anti-terminator regulator (ANTAR) family of response regulators, and its output domain controls gene expression by RNA binding (Shu & Zhulin, 2002). ANTAR proteins interact with nascent transcripts to prevent formation of transcriptional terminators that precede certain genes, allowing RNA polymerase to transit into the coding regions. The E. faecalis eut locus contains four of these rho-independent terminators, and significant evidence supports that they are the targets for antitermination by the eutV ANTAR protein (Fox, et al., 2009; Baker & Perego, 2011; Ramesh, et al., 2012). As already mentioned, expression of the genes downstream of the terminators is dependent on ethanolamine and the two-component system. Deletion of the terminators preceding eutP, eutS and eutG resulted in constitutive expression (Baker & Perego, 2011; Ramesh, et al., 2012). Finally, eutV binds to the RNA in a region just upstream and overlapping these terminators (Ramesh, et al., 2012). These data support an antitermination function for eutV and validates a model in which the eut locus is partially regulated by a series of rho-independent transcriptional terminators interspersed throughout the operon. A major focus of recent studies has been elucidating the details of the antiterminator structure that eutV recognizes.
Analysis of the sequences of the 5’ untranslated regions (UTR) of the E. faecalis eut genes revealed a hairpin structure with a short stem region and a hexanucleotide loop, with the first and fourth loop residues conserved among the E. faecalis eut genes (Ramesh, et al., 2012). A similar hairpin structure have been reported in the substrate of a previously characterized ANTAR protein, NasR from Klebsiella oxytoca (Chai & Stewart, 1999), and the loop sequences found to be conserved in the E. faecalis eut genes were shown to be important for the binding of NasR (Ramesh, et al., 2012; Chai & Stewart, 1999). A second, similar stem-loop structure was identified just downstream of the first hairpin, which also had a hexamer loop with similar conservation of loop residues. This second hairpin overlapped the 5’ end of the intrinsic terminator that precedes the ORF, such that formation of the hairpin and the terminator became mutually exclusive. This second stem-loop also contains a motif, called the ANTAR recognition sequence, that was predicted to be the site of recognition by eutV based on sequence conservation among eut genes present in Firmicutes (Chai & Stewart, 1999). This dual-hairpin motif and its conserved loop residues can be identified in the leaders of the eut genes from Clostridium species and Listeria monocytogenes, as well as in the substrates of the previously characterized ANTAR proteins, NasR and AmiR. This led to the hypothesis that upon activation, the eutV ANTAR protein bound the eut RNA, specifically interacting with the two terminal loops of the dual-hairpin motif in the nascent transcripts to stabilize an antiterminator structure and exclude terminator formation (Ramesh, et al., 2012; Aymerich & Steinmetz, 1992; Babitzke & Gollnick, 2001). This dual-hairpin motif is unique, as compared to the antiterminators employed by other bacterial regulators such as the Sac/Bgl and TRAP proteins (Aymerich & Steinmetz, 1992; Babitzke & Gollnick, 2001). In most previously characterized systems, the antiterminator is a single alternative structure that overlaps with the terminator in a manner such that only a single hairpin can exist.
Using a lacZ-based reporter assay, it was demonstrated that both stem-loops and the four conserved loop residues are critical for successful antitermination in vivo by eutV in the presence of ethanolamine and AdoCbl (Ramesh, et al., 2012). This was also supported by in vitro EMSA studies. In vivo data also showed that the terminal closing base pairs of the first hairpin may result in sequence-specific interactions with eutV. In addition, an optimal distance between the two stem-loops was required, and such an alteration resulted in reduced efficiency of antitermination. Hence, the current model for the mechanism of antitermination by the eutV ANTAR protein is formation of a protein-RNA complex involving the dual hairpin structure in the eut RNA, which prevents the formation of the terminator, resulting in expression of the downstream genes (Ramesh, et al., 2012).
The dual nature of the eut RNA substrate hinted at the possibility of interaction with a dimer of eutV (123). SEC-MALLS (Size Exclusion Chromatography – Multi-Angle Laser Light Scattering) analysis of full-length eutV indicated that it exists as a monomer in the unphosphorylated state. Phosphoryl transfer from the cognate sensor kinase, eutW, induced dimerization of eutV. DRaCALA (Differential radial capillary action of ligand assay) assays in turn showed that phospho-eutV bound the RNA substrate with significantly higher affinity, as compared to the unphosphorylated form of the protein. Interestingly, eutV that lacks the N-terminal domain is capable of dimerizing and binding the RNA substrate with higher affinity, as compared to unphosphorylated full-length protein, suggesting that the unphosphorylated receiver domain actively inhibits dimerization and therefore RNA binding. Overall, signal-induced dimerization of the eutV ANTAR protein seems to be essential for recognition of a symmetric, dual-hairpin RNA ligand, which is conceptually similar to the molecular mechanism exhibited by many DNA-binding response regulator proteins that dimerize and recognize paired sequences in the DNA. The E. faecalis eutV/eutW TCS provides, for the first time, a mechanistic model of antitermination employed by ANTAR proteins, validated by in vitro and in vivo techniques (Ramesh, et al., 2012).
An AdoCbl-Binding Riboswitch
The first step in the degradation of ethanolamine involves the EutBC ammonia lyase that converts ethanolamine into ammonia and acetaldehyde. AdoCbl is an essential co-factor of this enzyme (Roof & Roth, 1988; Roof & Roth, 1989; Scarlett & Turner, 1976). As a result, it is not surprising that AdoCbl also acts as a biochemical cue for the upregulation of the eut genes via an AdoCbl-binding riboswitch found in an intergenic region near the beginning of the locus (Figure 4) (Fox, et al., 2009; Baker & Perego, 2011). Riboswitches are non-coding, cis-acting RNA that serve as ligand-responsive genetic control elements that can modulate the expression of genes in response to changing concentrations of metabolites. They are usually present in the 5’ untranslated leader sequences that precede the coding region of genes. Riboswitches typically have two major domains: an aptamer region that binds the ligand with high specificity, and an expression platform that undergoes structural changes upon ligand-binding to control the expression of its target genes (Winkler & Breaker, 2005).
The E. faecalis eut locus has an AboCbl-binding riboswitch in the 5’UTR of the eutG gene (Fox, et al., 2009). A similar riboswitch has also been located in the eut locus of Listeria monocytogenes, which suggests conservation of this element. In-line probing studies have demonstrated that the eut riboswitch specifically binds AdoCbl with an affinity that is comparable to that of other AdoCbl riboswitches (Fox, et al., 2009). The affinity for cyanocobalamin is significantly poorer, demonstrating the specificity for AdoCbl. Gene control by riboswitches in prokaryotes involves control of either transcriptional termination or translation initiation. In Gram-positive bacteria, the binding of the ligand to its riboswitch typically results in stabilization of an intrinsic terminator, thereby shutting off gene expression (Winkler & Breaker, 2005). However, the eut riboswitch is unique, in that binding of AdoCbl to the aptamer region causes structural changes that prevent the formation of an intrinsic terminator that is located downstream of the riboswitch and upstream of the eutG gene (Fox, et al., 2009). In vitro transcription analysis showed increase in the runoff transcript concentration, with a greater amount of AdoCbl present in the reaction. Hence, the eut riboswitch appears to be a positively acting element that induces gene expression by causing antitermination at the terminator in front of eutG (Fox, et al., 2009). However, this terminator also has the conserved dual-stem loop motif suggestive of regulation by eutV, and it is not clear how both a riboswitch and an antiterminator protein influence formation of this single terminator.
A recent study by Baker and Perego suggested a different mechanism for the way in which the eut riboswitch acts (Baker & Perego, 2011). They identified a promoter region upstream of the eut riboswitch and a second promoter downstream of the riboswitch, but preceding the eutG ORF. Based on their data, they suggest that binding of AdoCbl to the riboswitch causes termination of the transcript that initiates from the first promoter within the riboswitch, rather than causing antitermination at the intrinsic terminator preceding eutG. They speculate that in the absence of AdoCbl, transcripts initiated from the first promoter continue through the second promoter. These read-through transcripts somehow prevent successful initiation at the second promoter. As a result, they suggest that binding of AdoCbl to the eut riboswitch facilitates the initiation of the second, more prevalent transcript by cutting off the interfering first transcript, and in this manner, the action of the AdoCbl riboswitch promotes downstream gene transcription (Baker & Perego, 2011).
Further analysis will be required to resolve the mechanism of action of the AdoCbl riboswitch, and whether or not interaction with AdoCbl promotes antitermination at the T2 terminator as Fox et al. propose; causes termination at a point earlier in the transcript as Baker et al. suggest; or functions differently than either model. Overall, the unique combination of features within the eut operon of E. faecalis makes it a particularly attractive model to investigate the mechanisms of post-transcription initiation regulation.
Conclusion
In discovering the ways in which E. faecalis responds to stimuli through transcriptional and post-transcriptional regulation, we begin to build an understanding of the complex mechanisms responsible for the success of the enterococci in adapting to a wide range of host environments. Response of E. faecalis to numerous effector signals, often relevant to the human host, have been explored, including nutrient availability, quorum signals, antibiotics, metal ions, bile levels, urine, and blood. Understanding the global transcriptional profile under these conditions provides insight into the dual personality of E. faecalis as both a commensal and pathogen.
Looking forward, a rapidly increased understanding of transcriptional regulation will inevitably be linked to next-generation sequencing technology, which may provide a new and improved method for mapping and quantification of microbial transcriptomes. As compared to the microarray-based technology, the sequence-based transcriptomics holds several advantages: (i) The transcriptome of an organism can be studied without any access to its genome sequence; (ii) the technology is not dependent on DNA sequenced probes; (iii) the dynamic range of the sequence technology is better than hybridization based technology; (iv) it is more sensitive and (v) the high throughput expression data are obtained by single nucleotide resolution. Moreover, next-generation sequencing also supplies information on regulatory RNA species as opposed to microarrays, thereby adding a new dimension to our understanding of microbial regulatory networks. Current efforts aimed at reducing the cost of sequencing by several orders of magnitude will further promote sequencing-based technologies, and will likely provide exponential growth in discoveries surrounding transcript regulation.
References
- Aakra A, Vebø H, Indahl U, Snipen L, Gjerstad O, Lunde M, et al. The Response of Enterococcus faecalis V583 to Chloramphenicol Treatment. Int J Microbiol. 2010;•••:483048. [PMC free article: PMC2902013] [PubMed: 20628561]
- Aakra Å, Vebø H, Snipen L, Hirt H, Aastveit A, Kapur V, et al. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob Agents Chemother. 2005;49(6):2246–2259. [PMC free article: PMC1140525] [PubMed: 15917518]
- Abrantes MC, Lopes Md, Kok J. Impact of manganese, copper and zinc ions on the transcriptome of the nosocomial pathogen Enterococcus faecalis V583. PLoS ONE. 2011;6(10):e26519. [PMC free article: PMC3203883] [PubMed: 22053193]
- Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Mol Biol. 2001;8:710–714. [PubMed: 11473263]
- Anderson JE, Ptashne M, Harrison SC. Structure of the repressor-operator complex of bacteriophage 434. Nature. 1987;326(6116):846–852. [PubMed: 3553959]
- Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179(1):97–106. [PMC free article: PMC178666] [PubMed: 8981985]
- Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174(8):2582–2591. [PMC free article: PMC205897] [PubMed: 1556077]
- Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci U S A. 1992;89(21):10410–10414. [PMC free article: PMC50348] [PubMed: 1279678]
- Babitzke P, Gollnick P. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J Bacteriol. 2001;183(20):5795–5802. [PMC free article: PMC99655] [PubMed: 11566976]
- Baker KA, Perego M. Transcription antitermination by a phosphorylated response regulator and cobalamin-dependent termination at a B₁₂ riboswitch contribute to ethanolamine utilization in Enterococcus faecalis. J Bacteriol. 2011;193(10):2575–2586. [PMC free article: PMC3133151] [PubMed: 21441515]
- Baptista M, Depardieu F, Reynolds P, Courvalin P, Arthur M. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol Microbiol. 1997;25(1):93–105. [PubMed: 11902729]
- Baptista M, Rodrigues P, Depardieu F, Courvalin P, Arthur M. Single-cell analysis of glycopeptide resistance gene expression in teicoplanin-resistant mutants of a VanB-type Enterococcus faecalis. Mol Microbiol. 1999;32(1):17–28. [PubMed: 10216856]
- Barcelona-Andrés B, Marina A, Rubio V. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J Bacteriol. 2002;184(22):6289–6300. [PMC free article: PMC151947] [PubMed: 12399499]
- Begley M, Sleator RD, Gahan CG, Hill C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun. 2005;73(2):894–904. [PMC free article: PMC546953] [PubMed: 15664931]
- Bitsori M, Maraki S, Raissaki M, Bakantaki A, Galanakis E. Community-acquired enterococcal urinary tract infections. Pediatr Nephrol. 2005;20(11):1583–1586. [PubMed: 15971072]
- Blancato VS, Repizo GD, Suárez CA, Magni C. Transcriptional regulation of the citrate gene cluster of Enterococcus faecalis Involves the GntR family transcriptional activator CitO. J Bacteriol. 2008;190(22):7419–7430. [PMC free article: PMC2576652] [PubMed: 18805984]
- Bøhle LA, Færgestad EM, Veiseth-Kent E, Steinmoen H, Nes IF, Eijsink VG, et al. Identification of proteins related to the stress response in Enterococcus faecalis V583 caused by bovine bile. Proteome Sci. 2010;8:37. [PMC free article: PMC2907315] [PubMed: 20579342]
- Booth MC, Bogie CP, Salh HG, Siezen RJ, Hatter KL, Gilmore MS. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol Microbiol. 1996;21(6):1175–1184. [PubMed: 8898386]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188(8):2875–2884. [PMC free article: PMC1446981] [PubMed: 16585749]
- Bourgogne A, Singh KV, Kox KA, Pflughoeft KJ, Murray BE, Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189(17):6490–6493. [PMC free article: PMC1951926] [PubMed: 17586623]
- Bourgogne A, Thomson LC, Murray BE. Bicarbonate enhances expression of the endocarditis and biofilm associated pilus locus, ebpR-ebpABC, in Enterococcus faecalis. BMC Microbiol. 2010;10:17. [PMC free article: PMC2824692] [PubMed: 20092636]
- Bowen R. (1998). Bile secretion and the enterohepatic circulation of bile acids. In F. Feldman, & M. Sleisenger (Eds.), Sleisenger and Fordtran's Gastrointestinal and Liver Disease (pp. 937-948). Philadelphia: W. B. Saunders, Co.
- Boyd DA, Willey BM, Fawcett D, Gillani N, Mulvey MR. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob Agents Chemother. 2008;52(7):2667–2672. [PMC free article: PMC2443935] [PubMed: 18458129]
- Breton YL, Mazé A, Harte A, Lemarinier S, Auffray Y, Rincé A. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr Microbiol. 2002;45(6):434–439. [PubMed: 12402085]
- Bron PA, Molenaar D, de Vos WM, Kleerebezem M. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J Appl Microbiol. 2006;100(4):728–738. [PubMed: 16553727]
- Chai W, Stewart V. RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J Mol Biol. 1999;292(2):203–216. [PubMed: 10493869]
- Cobine PA, Jones CE, Dameron CT. Role for zinc(II) in the copper(I) regulated protein CopY. J Inorg Biochem. 2002;88(2):192–196. [PubMed: 11803039]
- Coburn PS, Pillar CM, Jett BD, Haas W, Gilmore MS. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science. 2004;306(5705):2270–2272. [PubMed: 15618522]
- Coque TM, Patterson JE, Steckelberg JM, Murray BE. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J Infect Dis. 1995;171(5):1223–1229. [PubMed: 7751697]
- Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42 Suppl 1:S25–S34. [PubMed: 16323116]
- Del Papa MF, Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol. 2008;190(21):7147–7156. [PMC free article: PMC2580688] [PubMed: 18776017]
- Depardieu F, Bonora MG, Reynolds PE, Courvalin P. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol. 2003;50(3):931–948. [PubMed: 14617152]
- Depardieu F, Kolbert M, Pruul H, Bell J, Courvalin P. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2004;48(10):3892–3904. [PMC free article: PMC521886] [PubMed: 15388450]
- Depardieu F, Reynolds PE, Courvalin P. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob Agents Chemother. 2003;47(1):7–18. [PMC free article: PMC149003] [PubMed: 12499162]
- Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A. 2007;104(7):2384–2389. [PMC free article: PMC1892938] [PubMed: 17284603]
- Dobi A, Dameron CT, Hu S, Hamer D, Winge DR. Distinct regions of Cu(I).ACE1 contact two spatially resolved DNA major groove sites. J Biol Chem. 1995;270(17):10171–10178. [PubMed: 7730320]
- Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183(24):7341–7353. [PMC free article: PMC95583] [PubMed: 11717293]
- Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanS(B)-VanR (B) two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178(5):1302–1309. [PMC free article: PMC177803] [PubMed: 8631706]
- Forouhar F, Lee IS, Vujcic J, Shen J, Vorobiev SM, Xiao R, et al. Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase. J Biol Chem. 2005;280(48):40328–40336. [PubMed: 16210326]
- Foucault ML, Depardieu F, Courvalin P, Grillot-Courvalin C. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci U S A. 2010;107(39):16964–16969. [PMC free article: PMC2947908] [PubMed: 20833818]
- Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, et al. (2009). Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proceedings of the National Academy of Sciences(106), 4435-4440. [PMC free article: PMC2647976] [PubMed: 19246383]
- Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harey BR. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol. 2010;192(20):5489–5498. [PMC free article: PMC2950486] [PubMed: 20729365]
- Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. [PMC free article: PMC2950637] [PubMed: 20234377]
- Gravesen A, Ramnath M, Rechinger KB, Andersen N, Jänsch L, Héchard Y, et al. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology. 2002;148(Pt 8):2361–2369. [PubMed: 12177330]
- Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002;415(6867):84–87. [PubMed: 11780122]
- Hancock LE, Gilmore MS. (2002). The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proceedings of the National Academy of Sciences(99), 1574-1579. [PMC free article: PMC122232] [PubMed: 11830672]
- Hancock LE, Perego M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol. 2004;186(17):5629–5639. [PMC free article: PMC516840] [PubMed: 15317767]
- Handwerger S, Kolokathis A. Induction of vancomycin resistance in Enterococcus faecium by inhibition of transglycosylation. FEMS Microbiol Lett. 1990;58(2):167–170. [PubMed: 2227352]
- Héchard Y, Sahl HG. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie. 2002;84(5-6):545–547. [PubMed: 12423799]
- Holman TR, Wu Z, Wanner BL, Walsh CT. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry. 1994;33(15):4625–4631. [PubMed: 8161518]
- Huebner J, Wang Y, Krueger WA, Madoff LC, Martirosian G, Boisot S, et al. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect Immun. 1999;67(3):1213–1219. [PMC free article: PMC96449] [PubMed: 10024563]
- Jonas BM, Murray BE, Weinstock GM. Characterization of emeA, a NorA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob Agents Chemother. 2001;45(12):3574–3579. [PMC free article: PMC90871] [PubMed: 11709342]
- Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105(36):13580–13585. [PMC free article: PMC2533232] [PubMed: 18757757]
- Klare I, Werner G, Witte W. Enterococci. Habitats, infections, virulence factors, resistances to antibiotics, transfer of resistance determinants. Contrib Microbiol. 2001;8:108–122. [PubMed: 11764728]
- Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, et al. D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2011;55(10):4606–4612. [PMC free article: PMC3187002] [PubMed: 21807981]
- Lim D, Poole K, Strynadka NC. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem. 2002;277(32):29253–29259. [PubMed: 12034710]
- Linder MC, Hazegh-Azam M. Copper biochemistry and molecular biology. Am J Clin Nutr. 1996;63(5):797S–811S. [PubMed: 8615367]
- Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A. 1992;89(19):8938–8942. [PMC free article: PMC50039] [PubMed: 1409590]
- Lu ZH, Solioz M. Copper-induced proteolysis of the CopZ copper chaperone of Enterococcus hirae. J Biol Chem. 2001;276(51):47822–47827. [PubMed: 11585824]
- Lu ZH, Dameron CT, Solioz M. The Enterococcus hirae paradigm of copper homeostasis: copper chaperone turnover, interactions, and transactions. Biometals. 2003;16(1):137–143. [PubMed: 12572673]
- Lübben M, Portmann R, Kock G, Stoll R, Young MM, Solioz M. Structural model of the CopA copper ATPase of Enterococcus hirae based on chemical cross-linking. Biometals. 2009;22(2):363–375. [PubMed: 18979168]
- Malani PN, Kauffman CA, Zervis MJ. (2002). Enterococcal disease, epidemiology and treatment. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, & L. B. Rice (Eds.), The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance. Washington, D. C.: ASM Press.
- Marugg JD, Gonzalez CF, Kunka BS, Ledeboer AM, Pucci MJ, Toonen MY, et al. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58(8):2360–2367. [PMC free article: PMC195787] [PubMed: 1514784]
- Michaux C, Martini C, Hanin A, Auffray Y, Hartke A, Giard JC. SlyA regulator is involved in bile salts stress response of Enterococcus faecalis. FEMS Microbiol Lett. 2011;324(2):142–146. [PubMed: 22092815]
- Michaux C, Sanguinetti M, Reffuveille F, Auffray Y, Posteraro B, Gilmore MS, et al. SlyA is a transcriptional regulator involved in the virulence of Enterococcus faecalis. Infect Immun. 2011;79(7):2638–2645. [PMC free article: PMC3191995] [PubMed: 21536798]
- Mohamed JA, Tenq F, Nallapareddy SR, Murray BE. Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type i and fibronectin. J Infect Dis. 2006;193(2):231–240. [PubMed: 16362887]
- Nakayama J, Chen S, Oyama N, Nishiguchi K, Azab EA, Tanaka E, et al. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrd. J Bacteriol. 2006;188(23):8321–8326. [PMC free article: PMC1698201] [PubMed: 16980448]
- Nakayama J, Kariyama R, Kumon H. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl Environ Microbiol. 2002;68(6):3152–3155. [PMC free article: PMC123941] [PubMed: 12039782]
- Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, Erlandsen SL, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. [PMC free article: PMC1578622] [PubMed: 17016560]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. [PMC free article: PMC4313539] [PubMed: 19686078]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. [PubMed: 18713030]
- Odermatt A, Solioz M. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J Biol Chem. 1995;270(9):4349–4354. [PubMed: 7876197]
- Opsata M, Nes IF, Holo H. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 2010;10:224. [PMC free article: PMC2941500] [PubMed: 20738841]
- Patiño LA, Courvalin P, Perichon B. vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405. J Bacteriol. 2002;184(23):6457–6464. [PMC free article: PMC135418] [PubMed: 12426332]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. [PubMed: 12663927]
- Pechère JC. Macrolide resistance mechanisms in Gram-positive cocci. Int J Antimicrob Agents. 2001;18 Suppl 1:S25–S28. [PubMed: 11574191]
- Pfeiler EA, Azcarate-Peril MA, Klaenhammer TR. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J Bacteriol. 2007;189(13):4624–4634. [PMC free article: PMC1913432] [PubMed: 17449631]
- Pinkston KL, Gao P, Diaz-Garcia D, Sillanpää J, Nallapareddy SR, Murray BE, et al. The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J Bacteriol. 2011;193(17):4317–4325. [PMC free article: PMC3165527] [PubMed: 21705589]
- Portillo A, Ruiz-Larrea F, Zarazaga M, Alonso A, Martinez JL, Torres C. Macrolide resistance genes in Enterococcus spp. Antimicrob Agents Chemother. 2000;44(4):967–971. [PMC free article: PMC89799] [PubMed: 10722498]
- Qin X, Singh KV, Weinstock GM, Murray BE. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun. 2000;68(5):2579–2586. [PMC free article: PMC97462] [PubMed: 10768947]
- Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol. 2001;183(11):3372–3382. [PMC free article: PMC99635] [PubMed: 11344145]
- Ramesh A.DebRoy, S., Goodson, J. R., Kox, K. A., Faz, H., Garsin, D. A., et alThe mechanism for RNA recognition by ANTAR regulators of gene expression PLoS Genet 20128(6)e1002666. [PMC free article: PMC3369931] [PubMed: 22685413]
- Ramnath M, Beukes M, Tamura K, Hastings JW. Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin A-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl Environ Microbiol. 2000;66(7):3098–3101. [PMC free article: PMC92118] [PubMed: 10877813]
- Reyes A, Leiva A, Cambiazo V, Méndez MA, González M. Cop-like operon: structure and organization in species of the Lactobacillales order. Biol Res. 2006;39(1):87–93. [PubMed: 16629168]
- Reyes-Jara A, Latorre M, López G, Bourgogne A, Murray BE, Cambiazo V, et al. Genome-wide transcriptome analysis of the adaptive response of Enterococcus faecalis to copper exposure. Biometals. 2010;23(6):1105–1112. [PubMed: 20577782]
- Reynolds PE. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8(11):943–950. [PubMed: 2532132]
- Reynolds PE, Courvalin P. Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-D-serine. Antimicrob Agents Chemother. 2005;49(1):21–25. [PMC free article: PMC538887] [PubMed: 15616270]
- Ribeiro T, Santos S, Marques MI, Gilmore M, de Fátima Silva Lopes M. Identification of a new gene, vanV, in vanB operons of Enterococcus faecalis. Int J Antimicrob Agents. 2011;37(6):554–557. [PubMed: 21482081]
- Roberts JC, Singh KV, Okhuysen PC, Murray BE. Molecular epidemiology of the fsr locus and of gelatinase production among different subsets of Enterococcus faecalis isolates. J Clin Microbiol. 2004;42(5):2317–2320. [PMC free article: PMC404671] [PubMed: 15131223]
- Roberts MC. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2008;282(2):147–159. [PubMed: 18399991]
- Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170(9):3855–3863. [PMC free article: PMC211381] [PubMed: 3045078]
- Roof DM, Roth JR. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1989;171(6):3316–3323. [PMC free article: PMC210052] [PubMed: 2656649]
- Ruiz L, Zomer A, O'Connell-Motherway M, van Sinderen D, Margolles A. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl Environ Microbiol. 2012;78(4):1123–1131. [PMC free article: PMC3273026] [PubMed: 22156415]
- Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, et al. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33(9):1588–1591. [PMC free article: PMC172707] [PubMed: 2554802]
- Scarlett FA, Turner JM. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. Microbiology. 1976;95(1):173–176. [PubMed: 784902]
- Shepard BD, Gilmore MS. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect Immun. 2002;70(8):4344–4352. [PMC free article: PMC128128] [PubMed: 12117944]
- Shioya K, Michaux C, Kuenne C, Hain T, Verneuil N, Budin-Verneuil A, et al. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis V583. PLoS ONE. 2011;6(9):e23948. [PMC free article: PMC3166299] [PubMed: 21912655]
- Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002;27(1):3–5. [PubMed: 11796212]
- Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE, et al. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immun. 2002;70(10):5647–5650. [PMC free article: PMC128331] [PubMed: 12228293]
- Silva JC, Haldimann A, Prahalad MK, Walsh CT, Wanner BL. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc Natl Acad Sci U S A. 1998;95(20):11951–11956. [PMC free article: PMC21746] [PubMed: 9751771]
- Singh KV, Malathum K, Murray BE. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob Agents Chemother. 2001;45(1):263–266. [PMC free article: PMC90270] [PubMed: 11120975]
- Sleator RD, Hill C. Compatible solutes: the key to Listeria's success as a versatile gastrointestinal pathogen? Gut Pathogens. 2010;2:20. [PMC free article: PMC3006354] [PubMed: 21143981]
- Sleator RD, Wemekamp-Kamphuis HH, Gahan CG, Abee T, Hill C. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol. 2005;55(4):1183–1195. [PubMed: 15686563]
- Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, et al. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun. 2004;72(11):6373–6381. [PMC free article: PMC523057] [PubMed: 15501767]
- Solheim M, Aakra Å, Vebo H, Snipen L, Nes IF. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl Environ Microbiol. 2007;73(18):5767–5774. [PMC free article: PMC2074917] [PubMed: 17660310]
- Solioz M. Role of proteolysis in copper homoeostasis. Biochem Soc Trans. 2002;30(4):688–691. [PubMed: 12196165]
- Solioz M, Odermatt A. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J Biol Chem. 1995;270:9217–9221. [PubMed: 7721839]
- Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev. 2003;27:183–195. [PubMed: 12829267]
- Stock AM, Robinson VL, Goudreau PN. (200). Two-component signal transduction. Annu Rev Biochem. •••;69:183–215. [PubMed: 10966457]
- Storz H-A. (2000). Bacterial Stress Responses. Washington, D. C.: ASM Press.
- Strausak D, Solioz M. CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J Biol Chem. 1997;272(14):8932–8936. [PubMed: 9083014]
- Suárez CA, Blancato VS, Poncet S, Deutscher J, Magni C. CcpA represses the expression of the divergent cit operons of Enterococcus faecalis through multiple cre sites. BMC Microbiol. 2011;11:227. [PMC free article: PMC3198936] [PubMed: 21989394]
- Tasevska N, Runswick SA, McTaggart A, Bingham SA. Urinary sucrose and fructose as biomarkers for sugar consumption. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1287–1294. [PubMed: 15894688]
- Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179(8):2512–2518. [PMC free article: PMC178997] [PubMed: 9098046]
- Theilacker C, Kaczynski Z, Kropec A, Fabretti F, Sange T, Holst O, et al. Opsonic antibodies to Enterococcus faecalis strain 12030 are directed against lipoteichoic acid. Infect Immun. 2006;74(10):5703–5712. [PMC free article: PMC1594888] [PubMed: 16988246]
- Tsuda T, Toyoshima C. Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase. EMBO J. 2009;28(12):1782–1791. [PMC free article: PMC2699366] [PubMed: 19478797]
- Urbański NK, Beresewicz A. Generation of *OH initiated by interaction of Fe2+ and Cu+ with dioxygen; comparison with the Fenton chemistry. Acta Biochim Pol. 2000;47(4):951–962. [PubMed: 11996118]
- Vebø HC, Snipen L, Nes IF, Brede DA. The transcriptome of the nosocomial pathogen Enterococcus faecalis V583 reveals adaptive responses to growth in blood. PLoS ONE. 2009;4(11):e7660. [PMC free article: PMC2766626] [PubMed: 19888459]
- Vebø HC, Solheim M, Snipen L, Nes IF, Brede DA. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS ONE. 2010;5(8):e12489. [PMC free article: PMC2930860] [PubMed: 20824220]
- Vogel H, Altincicek B, Glöckner G, Vilcinskas A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics. 2011;12:308. [PMC free article: PMC3224240] [PubMed: 21663692]
- Whitehead K, Versalovic J, Roos S, Britton RA. Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl Environ Microbiol. 2008;74(6):1812–1819. [PMC free article: PMC2268311] [PubMed: 18245259]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. [PubMed: 16153177]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37(Database issue):D603–D610. [PMC free article: PMC2686599] [PubMed: 18953024]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. [PubMed: 15306996]
- Wright GD, Holman TR, Walsh CT. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993;32(19):5057–5063. [PubMed: 8494882]
- Wu RY, Zhang RG, Zagnitko O, Dementieva I, Maltzev N, Watson JD, et al. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J Biol Chem. 2003;278(22):20240–20244. [PMC free article: PMC2792031] [PubMed: 12649270]
- Wunderli-Ye H, Solioz M. Effects of promoter mutations on the in vivo regulation of the cop operon of Enterococcus hirae by copper(I) and copper(II). Biochem Biophys Res Commun. 1999;259(2):443–449. [PubMed: 10362527]
- Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother. 2010;54(11):4643–4647. [PMC free article: PMC2976141] [PubMed: 20733041]
- Xu Y, Murray BE, Weinstock GM. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66(9):4313–4323. [PMC free article: PMC108521] [PubMed: 9712783]
- Xu Y, Singh KV, Murray BE, Weinstock GM. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect Immun. 2000;68(2):815–823. [PMC free article: PMC97210] [PubMed: 10639451]
- Zhou PB, Thiele DJ. Isolation of a metal-activated transcription factor gene from Candida glabrata by complementation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991;88(14):6112–6116. [PMC free article: PMC52032] [PubMed: 2068090]
- Transcriptional and Post Transcriptional Control of Enterococcal Gene Regulation...Transcriptional and Post Transcriptional Control of Enterococcal Gene Regulation - Enterococci
Your browsing activity is empty.
Activity recording is turned off.
See more...
 7.
7.