Error status
RAPT is currently out of service. We are working on deploying a fix and apologize for the inconvenience.
Assemble and annotate your prokaryotic genomes with RAPT
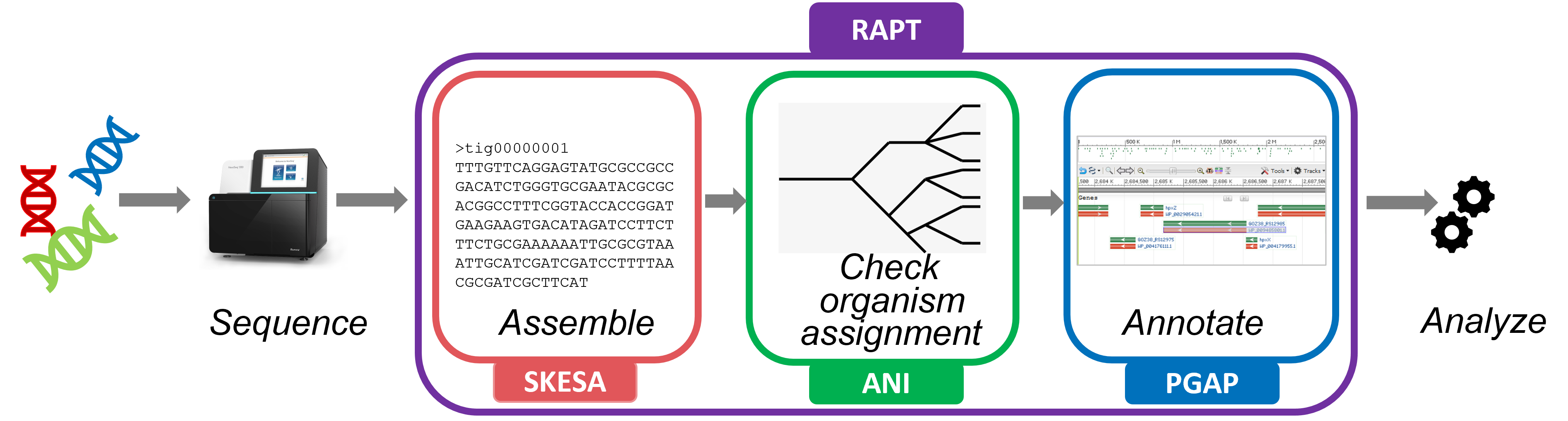
The Read assembly and Annotation Pipeline Tool (RAPT) is an easy-to-use pilot service for the de novo assembly and gene annotation of public or private Illumina genomic reads sequenced from bacterial or archaeal isolates. RAPT consists of three major components, the genome assembler SKESA, the taxonomic assignment tool ANI and the Prokaryotic Genome Annotation Pipeline (PGAP), and produces an annotated genome of quality comparable to RefSeq in a couple of hours (Learn more about RAPT).
With RAPT you will:
- assemble your reads into contigs
- assign a scientific name to the assembly
- predict coding and non-coding genes de novo, including anti-microbial resistance (AMR) genes and virulence factors, based on expert-curated data such as hidden Markov models and conserved domain architectures
- estimate the completeness and contamination level of the annotated assembly
RAPT Pipeline
Try it!
After you log in, provide an SRA accession, or upload a fastq file of reads and let it run! You will be notified by email that the job started, and a couple of hours later that the results are available for download.
Please visit the Documentation for more information about RAPT and the results it produces, and to learn how to run RAPT on your own machine.
We want to hear from you! RAPT is a NCBI Labs project and your Feedback is crucial in creating a user-driven service. After you use RAPT, we may contact you to hear more about your needs.
