- Exon count:
- 18
| Annotation release |
Status |
Assembly |
Chr
|
Location |
| RS_2024_08 |
current |
GRCh38.p14 (GCF_000001405.40) |
14 |
NC_000014.9 (92703809..92748627, complement) |
| RS_2024_08 |
current |
T2T-CHM13v2.0 (GCF_009914755.1) |
14 |
NC_060938.1 (86935604..86980392, complement) |
| RS_2024_09 |
previous assembly |
GRCh37.p13 (GCF_000001405.25) |
14 |
NC_000014.8 (93170154..93214972, complement) |
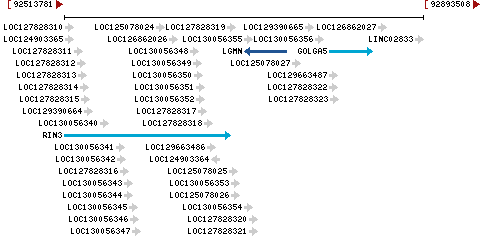
Chromosome 14 - NC_000014.9