dbVar Walkthrough
dbVar is NCBI's database of human genomic structural variation (SV). SV is defined as variation affecting 50 bp or more of DNA and includes insertions, deletions, duplications, inversions, multialleleic copy number changes, translocations, and complex variants. dbVar accepts submissions in Excel and VCF formats; variants with clinical assertions should be submitted instead to ClinVar, where they will be accessioned and then automatically propagated to dbVar.
This brief walkthrough demonstrates how dbVar can be a powerful tool to help with the clinical interpretation of novel structural variants. It highlights dbVar’s collection (nstd102) of nearly 60,000 clinical SV including clinical assertions (Benign, Likely benign, Uncertain significance, Likely pathogenic, Pathogenic) and demonstrates how to view these variants in the dbVar Genome Browser for any region in the genome.
The dbVar Genome Browser
You can navigate to the dbVar Genome Browser from the dbVar Homepage. (If you prefer, you can use NCBI's Genome Data Viewer GDV.)
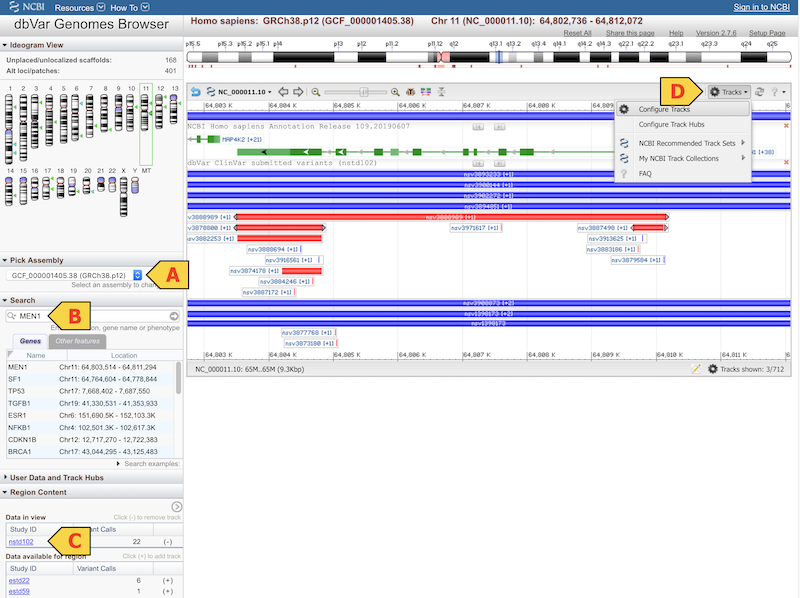
Choose an assembly (A)
Typically GRCh38 or GRCh37.
Search for a Gene, Location, or Phenotype (B)
In the Search pane to the left of the browser, enter any gene name, location, or phenotype, and press ‘return’. The browser will load the top result by default, as well as provide a list of additional results in the search pane for you to choose from.
Load the dbVar Clinical SV Collection (nstd102) (C)
If you are using the dbVar Genome Browser, navigate to the Region Content pane and find nstd102 among the studies listed in alphabetical order and click the plus ‘+’ sign to its right to add the track. If you are using GDV, navigate to the ‘Add Tracks’ pane, enter nstd102, and hit ‘return’.
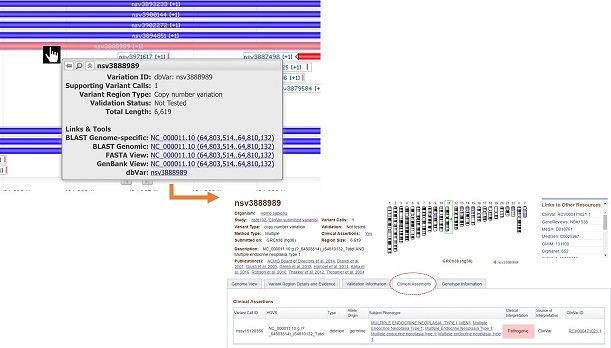
Individual variants can be investigated in more detail by mouse-over to reveal links to related records at NCBI. Click on the link to dbVar accession (ie. nsv3888989) to see variant details including clinical assertion.
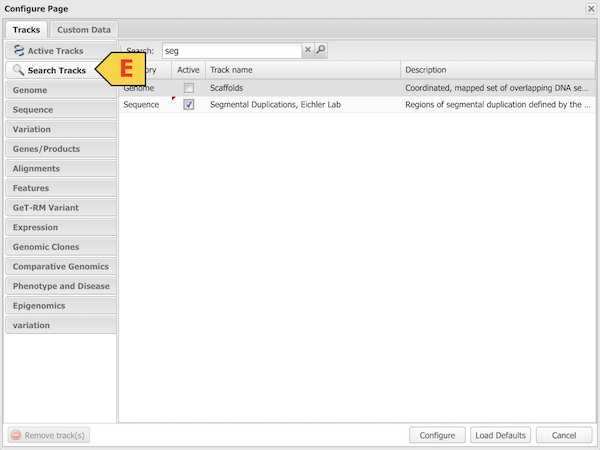
Configure Browser Tracks (D)
To hide unwanted tracks, click the small red ‘x’ in the top right corner of the track. To load additional tracks and configure the browser more extensively, use the Tracks pull-down (D) and select ‘Configure Tracks’. From the Configure Page (E, see below) you can search for and add other existing tracks as well as upload your own data. When you have adjusted settings the way you want, click ‘Configure’ and the browser will load the new information.
Additional Resources
Nonredundant Datasets (GitHub)
Please contact us if you have any questions or comments.
