Abstract
Many Streptococcus pyogenes strains harbor CRISPR-Cas loci that encode for the RNA-guided nuclease Cas9. Due to the use of this nuclease in genome editing, there has been a great deal of focus on the study of CRISPR-Cas immunity in these organisms, and as a result the S. pyogenes CRISPR-Cas9 system is one of the most studied and better understood of these systems. In this chapter, we review how the Cas9 nuclease mediates anti-phage immunity and how it can be repurposed for the genetic engineering of human cells and other eukaryotic organisms.
Introduction
Streptococcus pyogenes is the source of the most significant genetic tool of the twenty first century: the RNA-guided Cas9 nuclease (Pennisi, 2013). This nuclease is widely used to introduce genetic modifications in a variety of cells and organisms, from bacteria (Jiang, Bikard, Cox, Zhang, & Marraffini, 2013) and yeast (DiCarlo, et al., 2013) to monkeys (Niu, et al., 2014) and human cell lines (Cong, et al., 2013; Mali, et al., 2013a). The high efficiency and simplicity of the Cas9 genome editing technique has accelerated the possibilities of human gene therapy.
What is the function of Cas9 in Streptococcus pyogenes? The nuclease is a central player of the adaptive immunity that is provided by clustered regularly interspaced short palindromic repeats (CRISPR) loci (Marraffini, 2015). These loci consist of short repetitive sequences (30-40 bp) that are intercalated by equally short sequences of viral (bacteriophage) and plasmid origin (Bolotin, Quinguis, Sorokin, & Ehrlich, 2005; Mojica, Díez-Villaseñor, García-Martínez, & Soria, 2005; Pourcel, Salvignol, & Vergnaud, 2005) called “spacers.” The presence of a spacer sequence that matches the genome of a bacteriophage or conjugative plasmid prevents the host from becoming infected by these genetic invaders (Barrangou, et al., 2007; Marraffini & Sontheimer, 2008). Therefore, spacers provide sequence-specific immunity against bacteriophage and plasmid infection. Importantly, in a process known as adaptation, new spacers can be introduced into the CRISPR locus during infection (Barrangou, et al., 2007); in this way, the CRISPR system creates a memory of the infection that is used to provide immunity in subsequent encounters with the same invader or a related invader (for example, one that harbors the same spacer sequence in its genome) (Fig. 1A).
How do spacer sequences provide immunity? The CRISPR locus is usually transcribed into a long precursor RNA that contains both repeats and spacers. This precursor is subsequently cleaved at the repeat sequences to liberate small CRISPR RNAs (crRNAs) that contain the intervening spacer sequence (Tang, et al., 2002) (Fig. 1B). Cleavage is usually carried out by CRISPR-associated (Cas), repeat-specific endoribonucleases (Brouns, et al., 2008; Carte, Wang, Li, Terns, & Terns, 2008). The crRNA forms a ribonucleoprotein complex with an effector Cas nuclease, which finds its target (also known as the protospacer) through base-pairing of the spacer sequence in the crRNA and the genome of the invader, and proceeds to cleave it (Gasiunas, Barrangou, Horvath, & Siksnys, 2012; Hale, et al., 2009; Jinek, et al., 2012; Jore, et al., 2011; Samai, et al., 2015; Westra, et al., 2012) (Fig. 1C). The destruction of the viral or plasmid DNA stops the infection and confers immunity to the host (Garneau, et al., 2010). As expected from any host-pathogen interaction, this event is just a first step in the arms race between the host CRISPR-Cas systems and the extrachromosomal invaders, since bacteriophages and plasmids can evolve by generating mutations in the target site that prevent a perfect base-pairing with the spacer sequence of the crRNA (Deveau, et al., 2008). These mutations allow the invader to escape CRISPR-Cas immunity and re-establish infection. The cycle restarts when the CRISPR-Cas system acquires a new spacer sequence that matches perfectly with the genome of the invader (Levin, Moineau, Bushman, & Barrangou, 2013).
Streptococcus pyogenes CRISPR loci
Depending on the cas gene content, CRISPR-Cas systems can be classified into five types (Makarova, et al., 2015). Bioinformatic analysis revealed that seven of 13 available S. pyogenes genomes contain two CRISPR-Cas loci belonging to the types II (CRISPR-2) and 1 (CRISPR-1) (Nozawa, et al., 2011). Interestingly, the strains that lack CRISPR sequences have the highest number of prophages (Table 1, highlighted in green). More importantly, analysis of the CRISPR targets shows a mutually exclusive relationship between CRISPR spacer sequences and their prophage targets (Nozawa, et al., 2011; Marraffini L. A., 2010; Marraffini & Sontheimer, 2010). For example, strain SF370 has a total of 9 spacers, of which 6 match perfectly sequences found in 22 prophages present in other strains, and none matches any of the endogenous Φ370.1-4 prophages; the same is true for the other CRISPR-containing S. pyogenes strains (Table 1, highlighted in red). In addition, many spacers display 1-4 mismatches with prophage targets (within a 30-35 bp long spacer). Furthermore, in the M3 serotype MGAS315 and SSI-1 strains, the type II CRISPR-Cas locus is interrupted and inactivated by the prophages Φ315.1 and ΦSPsP5, respectively. The data suggests (i) that CRISPR immunity can prevent prophage acquisition in S. pyogenes; (ii) that phages have acquired mutations to evade CRISPR immunity and lysogenize; and (iii) that there is a dynamic relationship between S. pyogenes and its phages that results in the selection of strains with increased pathogenic adaptations. However, none of these hypotheses have been rigorously tested empirically, and the relationship between the CRISPR, prophages, and virulence properties of S. pyogenes strains still remains to be established. It also remains unclear whether CRISPR immunity prevents conjugation of the many different ICE elements present in S. pyogenes strains (Beres & Musser, 2007). Although CRISPR-Cas systems have been shown to prevent conjugation in other organisms, such as staphylococci (Marraffini & Sontheimer, 2008), no spacers have been found that match the conjugative elements of S. pyogenes.
Table 1.
CRISPR and prophage content of current S. pyogenes genomes.
Cas9-mediated defense
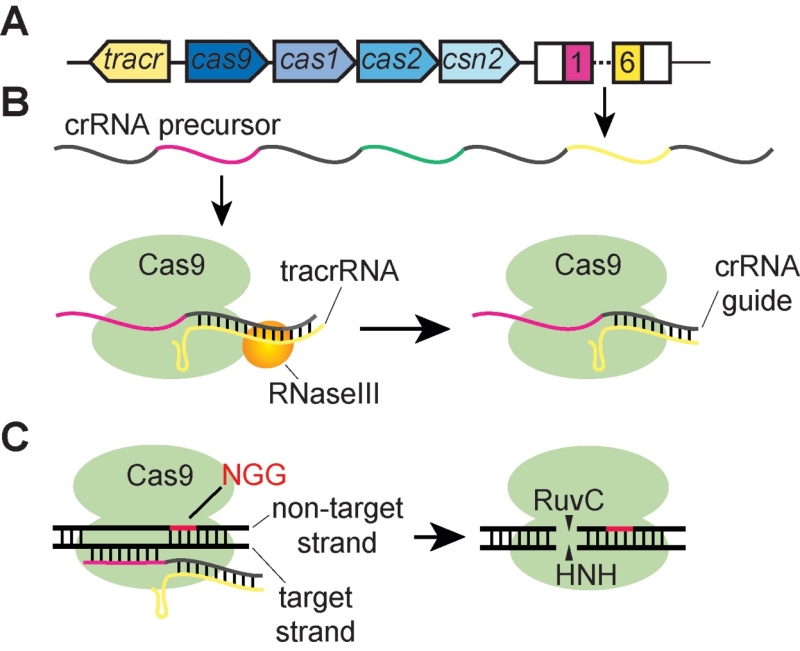
Although two CRISPR-Cas loci are present in many S. pyogenes strains (Nozawa, et al., 2011), it is not known whether the type I system is actually functional and/or if its cas genes and CRISPR sequences are expressed. In contrast, the molecular mechanism of S. pyogenes type II CRISPR-Cas immunity has been studied in detail. S. pyogenes SF370 contains a type IIA CRISPR-Cas system (Fig. 2A) that harbors four cas genes (cas9, cas1, cas2, and csn2), six 30-nt spacers flanked at each side by 36-nt repeats, and an additional gene encoding for a trans-encoded crRNA (tracrRNA) (Deltcheva, et al., 2011). Cas9 is central to the defense provided by the type II CRISPR Cas system, since it has been shown to be essential for all three stages of immunity (Fig. 1): adaptation (Heler, et al., 2015), crRNA biogenesis, (Deltcheva, et al., 2011) and interference (Sapranauskas, et al., 2011).
During the biogenesis of the crRNA guides, the CRISPR array of repeats and spacers is transcribed into a long precursor RNA. As opposed to the type I and III crRNA biogenesis pathways, the S. pyogenes type II systems do not require a repeat-specific endoribonuclease to process the crRNA precursor (Brouns, et al., 2008; Carte, Wang, Li, Terns, & Terns, 2008). Instead, the precursor is then processed by the combined action of the tracrRNA, RNase III, and Cas9 (Deltcheva, et al., 2011) (Fig. 2B). The tracrRNA contains an extensive secondary structure that is recognized and bound by Cas9. The tracrRNA also harbors a sequence that is complementary to the repeat sequence of the crRNA precursor. The annealing of these complementary sequences leads to the formation of a dsRNA that is cleaved at one end by RNase III. This cleavage liberates the small crRNAs from the precursor, which remain bound to Cas9 via their association with the tracrRNA. In this way, the processing of the type II crRNA precursor generates Cas9 molecules that are loaded with crRNA guides and ready to search invading DNA molecules for its targets. Although CRISPR immunity against phage infection has not been experimentally demonstrated in S. pyogenes, the S. pyogenes type II CRISPR system has been studied using vectors that were engineered to harbor prophage sequences that match the spacers contained in the CRISPR array (Deltcheva, et al., 2011). These experiments showed that the absence of the tracrRNA, RNase III, or Cas9 all prevent the destruction of the target-containing vector, a finding that is consistent with the essential role that these three elements play in the generation of crRNA guides.
Work performed in S. thermophilus determined that type II CRISPR-Cas immunity results in the introduction of double-strand DNA breaks (DSBs) on the genome of the invading phage or plasmid at the target site specified by spacer sequences (Garneau, et al., 2010). In addition, genetic analysis of the S. thermophilus CRISPR system demonstrated that cas9 is the only cas gene necessary for the interference phase of CRISPR immunity (Sapranauskas, et al., 2011). This mechanism presents the potential for an “autoimmune” nucleolytic reaction against the CRISPR locus itself; that is, how can Cas9 loaded with tracRNA and a crRNA avoid cleaving the spacer sequence from which the crRNA was transcribed? Early work in S. thermophilus revealed that the phage targets of the type II CRISPR-Cas system of this organism display a strong conservation of the sequence downstream of the target (Deveau, et al., 2008). The conserved nucleotides are referred to as the protospacer-adjacent motif or PAM (Mojica, Díez-Villaseñor, García-Martínez, & Almendros, 2009). The PAM is only present downstream of the target DNA, but is absent from the repeat sequences; that is, downstream of the spacer sequence in the CRISPR locus. Studies of the S. pyogenes type II CRISPR system confirmed the findings obtained for the S. thermophilus system: that cas9 is the only cas gene required for immunity (Heler, et al., 2015) and determined that the PAM motif sequence is NGG (Jiang, Bikard, Cox, Zhang, & Marraffini, 2013; Deltcheva, et al., 2011).
Biochemical and structural studies have meticulously characterized the crRNA-guide nucleolytic activity of Cas9 that is essential to provide immunity to the host (Fig. 2C). S. pyogenes Cas9 contains six domains: an HNH nuclease domain, a RuvC nuclease domain, an α-helical lobe, an arginine-rich region, a Topo-homology domain, and a PAM-recognition C-terminal domain (Jinek, et al., 2014; Nishimasu, et al., 2014). Single-molecule fluorescent experiments demonstrated that Cas9 searches for GG dinucleotide PAM sequences on the target dsDNA (Sternberg, Redding, Jinek, Greene, & Doudna, 2014). Transient binding to the PAM provides the necessary energy to unwind the dsDNA that is immediately upstream of the GG dinucleotide. Unwinding is followed by base-pairing of the crRNA with the seed sequence of the target. An inability to anneal leads to the quick release of Cas9, which continues sampling other DNA sequences. In contrast, if base pairing is productive, the rest of the crRNA sequence pairs with the target, forming an R-loop structure consisting of an RNA:DNA hybrid that is formed by the crRNA spacer sequence and its complementary DNA sequence (the target strand) and a displaced ssDNA (the PAM-containing, non-target strand) (Jiang, et al., 2016) (Fig. 2C). The formation of the R-loop triggers the cleavage of both DNA strands, with the HNH domain cleaving the target strand and the RuvC domain cleaving the PAM-containing strand (Jinek, et al., 2012; Jiang, et al., 2016).
Generation of a memory of infection
In contrast to crRNA biogenesis and Cas9 nuclease activity, the acquisition of new spacers following phage infection by the type IIA CRISPR-Cas system of S. pyogenes is less well understood. The original observation of spacer acquisition was done in Streptococcus thermophilus, where lytic phages were added to liquid cultures and phage-resistant bacteria were isolated (Barrangou, et al., 2007). The study showed that many of the bacteriophage-insensitive mutants (BIMs) expanded the CRISPR array by the incorporation of a new spacer with a sequence matching the genome of the infecting phage. However, studies like this are difficult to carry out in S. pyogenes, due to the lack of potent lytic phages that enable the isolation of BIMs. To overcome this limitation, a recent study transplanted the type IIA CRISPR-Cas system from S. pyogenes SF370 to Staphylococcus aureus (Heler, et al., 2015). Using staphylococcal lytic phages, the authors were able to observe and study the acquisition of new spacers in this CRISPR-Cas system. This arrangement was used to investigate the sampling of spacer sequences with functional flanking PAM sequences. Two scenarios are possible: (i) only spacers with the required NGG flanking nucleotide sequence are acquired; (ii) any phage sequence can become a new spacer, but only those flanked by NGG PAMs can provide immunity, and therefore are selected during phage infection. Genetic and biochemical analysis demonstrated the first scenario, with the PAM-binding domain of Cas9 being required during spacer acquisition to determine phage sequences that are flanked by a correct PAM (Heler, et al., 2015). This is a simple mechanism in which the same enzyme that is responsible for the recognition of the PAM during the implementation of immunity, Cas9, is also used to ensure that new spacers are flanked by such sequences.
The next step after the selection of a 30-bp phage sequence flanked by NGG is to integrate this sequence into the CRISPR array as a new spacer. This has not been investigated in S. pyogenes, but experiments with Escherichia coli demonstrated that Cas1 and Cas2 form a complex that is necessary and sufficient to integrate new spacers (Arslan, Hermanns, Wurm, Wagner, & Pul, 2014; Nuñez, et al., 2014; Nuñez, Lee, Engelman, & Doudna, 2015). Genetic analysis demonstrated that all four cas genes (cas9, cas1, cas2, and csn2) of the S. pyogenes system are necessary for spacer acquisition (Heler, et al., 2015). Altogether, these results suggest that Cas1 and Cas2 also perform spacer integration in S. pyogenes—possibly through a more elaborate mechanism than that of the type I E. coli CRISPR-Cas system, since Csn2 and Cas9 (which are present in the type II CRISPR-Cas system from S. pyogenes, but not in the type I system of E. coli) also seem to form a complex with Cas1 and Cas2 (Heler, et al., 2015). As explained above, the function of Cas9 in this complex is presumably to sample PAM sequences, but the function of Csn2 is unknown (Arslan, et al., 2013).
Cas9-based genetic applications
Since the examination of the crRNA-guided DNA targeting mechanism (Garneau, et al., 2010; Marraffini, 2010), Cas nucleases have been proposed as useful biotechnological tools that require the sequence-specific cleavage of DNA (Marraffini & Sontheimer, 2008; Sontheimer & Marraffini, 2010). One such application is the genetic engineering of eukaryotic cells. Early work with the yeast Saccharomyces cerevisiae demonstrated that the introduction of DSBs results in the generation of indels at the cleavage site after repair of the break via non-homologous end joining (NHEJ) (Plessis, Perrin, Haber, & Dujon, 1992; Rudin, Sugarman, & Haber, 1989). In addition, if the appropriate DNA template is provided after chromosomal cleavage, it could be used by the homology-directed repair (HDR) mechanism. Through this method, specific point mutations contained in the repair template can be introduced into the genome (Choulika, Perrin, Dujon, & Nicolas, 1995; Rouet, Smih, & Jasin, 1994). Therefore, a central aspect of this technique is the generation of a DSB at the desired sequence; however, the tools to achieve this have been difficult to engineer. Sequence-specific nucleases, such as zinc-finger (Bibikova, et al., 2001) and TALE nucleases (Christian, et al., 2010) were developed for this purpose, but programming them to achieve sequence-specific cleavage is difficult. In contrast, S. pyogenes Cas9 provides a simple and robust tool for the generation of precise DSBs, since the specificity can be easily programmed with the crRNA guide (Fig. 3). To make the system more amenable to genome editing, the tracrRNA and crRNA can be fused into a single-guide RNA (sgRNA) (Jinek, et al., 2012), thus reducing the number of components that need to be transferred from S. pyogenes to the target organism from three (tracrRNA, crRNA, and Cas9) to two (sgRNA and Cas9).
Genome editing of human cells mediated by S. pyogenes Cas9 was first achieved by transfecting a vector that harbored the sgRNA and the cas9 genes into HK293 cells to generate indels in the EMX1 chromosomal gene (Cong, et al., 2013) or in a gfp reporter gene (Mali, et al., 2013b) through the NHEJ pathway. In addition, it was shown that using an appropriate template for HDR-specific point mutations can be introduced into the EMX1 gene (Cong, et al., 2013). For this, the expression of cas9 was driven by the elongation factor 1α (EF1 α) promoter, its codons were changed for optimal translation in human cells, and nuclear localization signals were added to direct the nuclease to the cell nucleus. Gene knock-out through NHEJ-mediated indel generation is highly efficient and therefore can also be performed using libraries of sgRNAs that target the whole human genome to facilitate forward genetic experiments (Shalem, et al., 2014; Wang, Wei, Sabatini, & Lander, 2014). In this approach, a library of lentiviral vectors carrying cas9 and multiple sgRNAs is used to infect cells. Upon integration of the lentiviral vector into the human genome, Cas9 introduces a sgRNA-specific indel, with each cell expressing a different sgRNA and therefore having a different gene knock-out. This genetically heterogeneous cell population can be subjected to different selection pressures that favor or disfavor different genotypes. Next-generation sequencing of the lentiviral sgRNA locus of the cells under selection allows the identification of the gene knock-outs that are enriched or depleted, thus assigning the responsibility for particular phenotypes to specific genes. Other versions of the Cas9 technology were developed by the direct injection of cas9 mRNA and sgRNA molecules (Wang, et al., 2013), or of the Cas9 nuclease loaded with a sgRNA (Sung, et al., 2014). These techniques have been implemented to mutate multiple organisms, including mice (Wang, et al., 2013), flies (Gratz, et al., 2013), worms (Friedland, et al., 2013), livestock (Tan, et al., 2013), monkeys (Niu, et al., 2014), and many more. Moreover, the potential exists to use the technology to mutate the human germline: an outcome with important ethical consequences that requires serious study (Baltimore, et al., 2015).
The simplicity of the Cas9 DNA recognition mechanism has been successfully exploited to develop other applications besides genome editing. The cleavage of bacterial chromosomal sequences by S. pyogenes Cas9 is lethal (Bikard, Hatoum-Aslan, Mucida, & Marraffini, 2012), presumably because most bacteria lack the ability for NHEJ repair (Shuman & Glickman, 2007), and the nuclease repeatedly cleaves the target every time it is repaired by HDR. This lethality has been exploited to select for bacteria that carry mutations to prevent Cas9 cleavage (such as in the PAM or seed sequences) and thus enhance bacterial mutagenesis protocols (Jiang, Bikard, Cox, Zhang, & Marraffini, 2013), as well as to develop sequence-specific antimicrobials (Bikard, et al., 2014; Citorik, Mimee, & Lu, 2014). In addition, S. pyogenes Cas9 can be converted into an RNA-guided dsDNA binding protein if key residues are mutated in each of the nucleolytic active sites (D10A in the RuvC domain; H840A in the HNH domain). This “dead” protein, or dCas9, can be fused to different functional domains to bring them to specific sequences of the human genome. For example, the binding to promoter sequences of dCas9 fused to transcription activators or repressors can be used to modulate gene expression in human cells (Qi, et al., 2013). Similarly, the binding of fusion proteins that consist of dCas9 and chromatin modification enzymes can lead to the modification of nucleosomal histones for the silencing or activation of particular chromosomal regions (Hilton, et al., 2015; Kearns, et al., 2015). In addition, dCas9-Gfp fusions can be used to fluorescently mark different loci (Chen, et al., 2013).
Conclusions
Although the diseases caused by S. pyogenes produce a great deal of human suffering, this organism is also the source of Cas9, a nuclease that holds an enormous promise for both human genome editing and gene therapy. Ironically, an organism responsible for some of the most prevalent infectious diseases in the world could harbor a cure for a number of genetic diseases. This irony highlights the importance of the research being performed with S. pyogenes and other bacterial pathogens, which may not only provide new therapies to combat disease, but also could lead to new genetic tools that can revolutionize medicine. Because of the revolution in human genetics caused by Cas9, much of the recent research on the CRISPR-Cas system of S. pyogenes has been centered on the biochemistry and structure of this nuclease. Future work will address the function of the CRISPR-Cas locus in the ecology, evolution and pathogenesis of S. pyogenes.
Acknowledgments
The author is supported by the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award, a Burroughs Wellcome Fund PATH award, and a NIH Director’s New Innovator Award (1DP2AI104556-01).
Abbreviations
BIMs, bacteriophage-insensitive mutants
Cas, CRISPR-associated protein
CRISPR, clustered regularly interspaced short palindromic repeats
crRNA, CRISPR RNA
DSBs, double-strand DNA breaks
HDR, homology directed repair
ICE, integrated chromosomal element
NGG, PAM sequence for S. pyogenes Cas9
NHEJ, non-homologous end joining
PAM, protospacer-adjacent motif
sgRNA, single-guide RNA
TALE, transcription activator-like effector
tracrRNA, trans-encoded crRNA
ZFN, Zinc-finger nuclease
References
- Arslan Z., Hermanns V., Wurm R., Wagner R., Pul Ü. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Research. 2014;42(12):7884–7893. [PMC free article: PMC4081107] [PubMed: 24920831]
- Arslan Z., Wurm R., Brener O., Ellinger P., Nagel-Steger L., Oesterhelt F., et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Research. 2013;41(12):6347–6359. [PMC free article: PMC3695520] [PubMed: 23625968]
- Baltimore D., Berg P., Botchan M., Carroll D., Charo R. A., Church G., et al. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348(6230):36–38. [PMC free article: PMC4394183] [PubMed: 25791083]
- Banks D. J., Porcella S. F., Barbian K. D., Beres S. B., Philips L. E., Voyich J. M., et al. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. The Journal of Infectious Diseases. 2004;190(4):727–738. [PubMed: 15272401]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. [PubMed: 17379808]
- Beres S. B., Musser J. M. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PloS One. 2007;2(8):e800. [PMC free article: PMC1949102] [PubMed: 17726530]
- Beres S. B., Richter E. W., Nagiec M. J., Sumby P., Porcella S. F., DeLeo F. R., et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(18):7059–7064. [PMC free article: PMC1459018] [PubMed: 16636287]
- Beres S. B., Sylva G. L., Barbian K. D., Lei B., Hoff J. S., Mammarella N. D., et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(15):10078–10083. [PMC free article: PMC126627] [PubMed: 12122206]
- Bibikova M., Carroll D., Segal D. J., Trautman J. K., Smith J., Kim Y. G., et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Molecular and Cellular Biology. 2001;21(1):289–297. [PMC free article: PMC88802] [PubMed: 11113203]
- Bikard D., Euler C. W., Jiang W., Nussenzweig P. M., Goldberg G. W., Duportet X., et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature Biotechnology. 2014;32:1146–1150. [PMC free article: PMC4317352] [PubMed: 25282355]
- Bikard D., Hatoum-Aslan A., Mucida D., Marraffini L. A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host and Microbe. 2012;12(2):177–186. [PubMed: 22901538]
- Bolotin A., Quinguis B., Sorokin A., Ehrlich S. D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(Pt 8):2551–2561. [PubMed: 16079334]
- Brouns S. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., Snijders A. P., et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. [PMC free article: PMC5898235] [PubMed: 18703739]
- Carte J., Wang R., Li H., Terns R. M., Terns M. P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes and Development. 2008;22(24):3489–3496. [PMC free article: PMC2607076] [PubMed: 19141480]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., Li G. W., et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491. [PMC free article: PMC3918502] [PubMed: 24360272]
- Choulika A., Perrin A., Dujon B., Nicolas J. F. Induction of Homologous Recombination in Mammalian Chromosomes by Using the I-Scei System of Saccharomyces-Cerevisiae. Molecular and Cellular Biology. 1995;15(4):1968–1973. [PMC free article: PMC230423] [PubMed: 7891691]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., Hummel A., et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. [PMC free article: PMC2942870] [PubMed: 20660643]
- Citorik R. J., Mimee M., Lu T. K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature Biotechnology. 2014;32:1141–1145. [PMC free article: PMC4237163] [PubMed: 25240928]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339(6121):819–823. [PMC free article: PMC3795411] [PubMed: 23287718]
- Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. [PMC free article: PMC3070239] [PubMed: 21455174]
- Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. Journal of Bacteriology. 2008;190(4):1390–1400. [PMC free article: PMC2238228] [PubMed: 18065545]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., Church G. M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Research. 2013;41(7):4336–4343. [PMC free article: PMC3627607] [PubMed: 23460208]
- Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(8):4658–4663. [PMC free article: PMC31890] [PubMed: 11296296]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., Calarco J. A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Methods. 2013;10:741–743. [PMC free article: PMC3822328] [PubMed: 23817069]
- Garneau J. E., Dupuis M.-È., Villion M., Romero D. A., Barrangou R., Boyaval P., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. [PubMed: 21048762]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(39):E2579–E2586. [PMC free article: PMC3465414] [PubMed: 22949671]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., Harrison M. M., et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. [PMC free article: PMC3730909] [PubMed: 23709638]
- Green N. M., Zhang S., Porcella S. F., Nagiec M. J., Barbian K. D., Beres S. B., et al. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. The Journal of Infectious Diseases. 2005;192(5):760–770. [PubMed: 16088825]
- Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., Wells L., et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–956. [PMC free article: PMC2951265] [PubMed: 19945378]
- Heler R., Samai P., Modell J. W., Weiner C., Goldberg G. W., Bikard D., et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. [PMC free article: PMC4385744] [PubMed: 25707807]
- Hilton I. B., D'Ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnology. 2015;33(5):510–517. [PMC free article: PMC4430400] [PubMed: 25849900]
- Holden M. T., Scott A., Cherevach I., Chillingworth T., Churcher C., Cronin A., et al. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain manfredo. Journal of Bacteriology. 2007;189(4):1473–1477. [PMC free article: PMC1797351] [PubMed: 17012393]
- Jiang F., Taylor D. W., Chen J. S., Kornfeld J. E., Zhou K., Thompson A. J., et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. [PMC free article: PMC5111852] [PubMed: 26841432]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology. 2013;31:233–239. [PMC free article: PMC3748948] [PubMed: 23360965]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [PMC free article: PMC6286148] [PubMed: 22745249]
- Jinek M., Jiang F., Taylor D. W., Sternberg S. H., Kaya E., Ma E., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. [PMC free article: PMC4184034] [PubMed: 24505130]
- Jore M. M., Lundgren M., van Duijn E., Bultema J. B., Westra E. R., Waghmare S. P., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Structural & Molecular Biology. 2011;18:529–536. [PubMed: 21460843]
- Kearns N. A., Pham H., Tabak B., Genga R. M., Silverstein N. J., Garber M., et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nature Methods. 2015;12(5):401–403. [PMC free article: PMC4414811] [PubMed: 25775043]
- Levin B. R., Moineau S., Bushman M., Barrangou R. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genetics. 2013;9(3):e1003312. [PMC free article: PMC3597502] [PubMed: 23516369]
- Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., et al. An updated evolutionary classification of CRISPR-Cas systems. Nature Reviews Microbiology. 2015;13:722–736. [PMC free article: PMC5426118] [PubMed: 26411297]
- Mali P., Aach J., Stranges B., Esvelt K. M., Moosburner M., Kosuri S., et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology. 2013a;31:833–838. [PMC free article: PMC3818127] [PubMed: 23907171]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013b;339(6121):823–826. [PMC free article: PMC3712628] [PubMed: 23287722]
- Marraffini L. A. Impact of CRISPR immunity on the emergence of bacterial pathogens. Future Microbiology. 2010;5:693–695. [PubMed: 20441541]
- Marraffini L. A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526(7571):55–61. [PubMed: 26432244]
- Marraffini L. A., Sontheimer E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. [PMC free article: PMC2695655] [PubMed: 19095942]
- Marraffini L. A., Sontheimer E. J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Reviews Genetics. 2010;11:181–190. [PMC free article: PMC2928866] [PubMed: 20125085]
- McShan W. M., Ferretti J. J., Karasawa T., Suvorov A. N., Lin S., Qin B., et al. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. Journal of Bacteriology. 2008;190(23):7773–7785. [PMC free article: PMC2583620] [PubMed: 18820018]
- Mojica F. J., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. [PubMed: 19246744]
- Mojica F. J., Díez-Villaseñor C., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution. 2005;60(2):174–182. [PubMed: 15791728]
- Nakagawa I., Kurokawa K., Yamashita A., Nakata M., Tomiyasu Y., Okahashi N., et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Research. 2003;13(6A):1042–1055. [PMC free article: PMC403657] [PubMed: 12799345]
- Nishimasu H., Ran F. A., Hsu P. D., Konermann S., Shehata S. I., Dohmae N., et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. [PMC free article: PMC4139937] [PubMed: 24529477]
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–843. [PubMed: 24486104]
- Nozawa T., Furukawa N., Aikawa C., Watanabe T., Haobam B., Kurokawa K., et al. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One. 2011;6(5):e19543. [PMC free article: PMC3089615] [PubMed: 21573110]
- Nuñez J. K., Kranzusch P. J., Noeske J., Wright A. V., Davies C. W., Doudna J. A. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nature Structural and Molecular Biology. 2014;21:528–534. [PMC free article: PMC4075942] [PubMed: 24793649]
- Nuñez J. K., Lee A. S., Engelman A., Doudna J. A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. [PMC free article: PMC4359072] [PubMed: 25707795]
- Pennisi E. The CRISPR craze. Science. 2013;341(6148):833–836. [PubMed: 23970676]
- Plessis A., Perrin A., Haber J. E., Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130(3):451–460. [PMC free article: PMC1204864] [PubMed: 1551570]
- Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151(Pt 3):653–663. [PubMed: 15758212]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. [PMC free article: PMC3664290] [PubMed: 23452860]
- Rouet P., Smih F., Jasin M. Introduction of Double-Strand Breaks into the Genome of Mouse Cells by Expression of a Rare-Cutting Endonuclease. Molecular and Cellular Biology. 1994;14(12):8096–8106. [PMC free article: PMC359348] [PubMed: 7969147]
- Rudin N., Sugarman E., Haber J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122(3):519–534. [PMC free article: PMC1203726] [PubMed: 2668114]
- Samai P., Pyenson N., Jiang W., Goldberg G. W., Hatoum-Aslan A., Marraffini L. A. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161(5):1164–1174. [PMC free article: PMC4594840] [PubMed: 25959775]
- Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research. 2011;39(21):9275–9282. [PMC free article: PMC3241640] [PubMed: 21813460]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343(6166):84–87. [PMC free article: PMC4089965] [PubMed: 24336571]
- Shuman S., Glickman M. S. Bacterial DNA repair by non-homologous end joining. Nature Reviews Microbiology. 2007;5(11):852–861. [PubMed: 17938628]
- Smoot J. C., Barbian K. D., Van Gompel J. J., Smoot L. M., Chaussee M. S., Sylva G. L., et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4668–4673. [PMC free article: PMC123705] [PubMed: 11917108]
- Sontheimer E. J., Marraffini L. A. Microbiology: slicer for DNA. Nature. 2010;468:45–46. [PMC free article: PMC3045673] [PubMed: 21048757]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. [PMC free article: PMC4106473] [PubMed: 24476820]
- Sumby P., Porcella S. F., Madrigal A. G., Barbian K. D., Virtaneva K., Ricklefs S. M., et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. The Journal of Infectious Diseases. 2005;192(5):771–782. [PubMed: 16088826]
- Sung Y. H., Kim J. M., Kim H.-T., Lee J., Jeon J., Jin Y., et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Research. 2014;24:125–131. [PMC free article: PMC3875853] [PubMed: 24253447]
- Tan W., Carlson D. F., Lancto C. A., Garbe J. R., Webster D. A., Hackett P. B., et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16526–16531. [PMC free article: PMC3799378] [PubMed: 24014591]
- Tang T. H., Bachellerie J. P., Rozhdestvensky T., Bortolin M. L., Huber H., Drungowski M., et al. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(11):7536–7541. [PMC free article: PMC124276] [PubMed: 12032318]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153(4):910–918. [PMC free article: PMC3969854] [PubMed: 23643243]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S. Genetic Screens in Human Cells Using the CRISPR/Cas9 System. Science. 2014;343(6166):80–84. [PMC free article: PMC3972032] [PubMed: 24336569]
- Westra E. R., van Erp P. B., Künne T., Wong S. P., Staals R. H., Seegers C. L., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Molecular Cell. 2012;46(5):595–605. [PMC free article: PMC3372689] [PubMed: 22521689]
Publication Details
Author Information and Affiliations
Publication History
Created: April 7, 2016.
Copyright
Except where otherwise noted, this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC-BY-NC-ND 4.0). To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
Publisher
University of Oklahoma Health Sciences Center, Oklahoma City (OK)
NLM Citation
Marraffini LA. The CRISPR-Cas system of Streptococcus pyogenes: function and applications. 2016 Apr 7. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes : Basic Biology to Clinical Manifestations [Internet]. Oklahoma City (OK): University of Oklahoma Health Sciences Center; 2016-.