NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Two-pore domain potassium channels play important roles in regulation of cell membrane potential. Their activities modulate a variety of physiological processes including immune response, hormone secretion, chemosensation, and neuronal function. ML365 was identified as a novel selective small molecule inhibitor of the TASK1 or potassium channel, subfamily K, member 9 (KCNK3) two-pore domain potassium channel following a high throughput fluorescent screen of the Molecular Libraries Small Molecule Repository (MLSMR) library and structure activity relationship (SAR) analysis of active compounds. The fluorescent screen measuring thallium influx through TASK1 channels was used to identify the bisamide class of inhibitors. Chemical modification yielded a potent and selective inhibitor, ML365. The compound blocks TASK1 channels in both the thallium influx fluorescent assay (IC50 = 4 nM) and an automated electrophysiology assay (IC50 = 16 nM). Based on potency differences, it possesses more than 60-fold selectivity for inhibition of TASK1 over a closely-related, two-pore domain potassium channel, TASK3. ML365 displays little or no inhibition at 30 μM of more distantly related potassium channels, Kir2.1, potassium voltage-gated channel, KQT-like subfamily, member 2 (KCNQ2), and human ether-a go-go-related gene (hERG). Based on these criteria, ML365 is a best-in-class probe and is a useful pharmacological probe for in vitro studies of TASK1 function and in further studies aimed at developing therapeutic intervention.
Assigned Assay Grant #: 1 R03 MH090849-01
Screening Center Name & PI: Johns Hopkins Ion Channel Center, Min Li
Chemistry Center Name & PI: University of Kansas Specialized Chemistry Center, Jeffrey Aubé
Assay Submitter & Institution: Min Li, Ph.D., Johns Hopkins University, School of Medicine
PubChem Summary Bioassay Identifier (AID): 602490
Probe Structure & Characteristics

Table 1Summary of ML365 pharmacology†
| CID / ML# | Target Name | TASK1 (KCNK3) thallium flux (FDSS) assay** IC50 (nM) [SID 104221970, AID 652212] | Anti-target Names | Anti-target percent inhibition at 30 μM | Anti-target thallium assay IC50 (μM) [SID 104221970, AID 652190, AID 652194, AID 652192, AID 652211, AID 652188]] | Thallium assay flux (FDSS) Fold Selectivity* | TASK1 (KCNK3) QPatch: IC50 (nM) [SID 104221970, AID 652209] | TASK3 (KCNK9) QPatch: IC50 (nM) [SID 104221970, AID 652210] | QPatch Fold Selectivity* |
|---|---|---|---|---|---|---|---|---|---|
| CID 46785920 ML365 | KCNK3 or TASK1 | 4 nM | Kir2.1 | 2.1 | > 30 | > 7500 | 16 nM | 990 nM | 62 |
| hERG | 9.5 | > 30 | > 7500 | ||||||
| KCNQ2 | 42.4 | > 30 | > 7500 | ||||||
| TASK3 (KCNK9) | 91.9 | 0.39 | 98 | ||||||
| TASK1 (KCNK3) non-induced (parental) | 2.7 | > 30 | > 7500 |
- †
ML365 data shown is averaged from multiple experiments
- *
Selectivity = anti-target IC50/TASK1 IC50
- **
Assay concentration done at 3 μM; TASK1 percent inhibition was 96.6%
1. Recommendations for Scientific Use of the Probe
TWIK-related acid-sensitive potassium (TASK) channels belong to the two-pore domain potassium channel family. TASK1 and TASK3, two TASK channels, regulate cell membrane potential and thereby play important roles in regulating a variety of physiological processes. Biophysical experiments and studies with knockout animals have suggested specific roles of TASK1 and TASK3 in the activation and effector functions of T lymphocytes, cerebellum granual neuron viability, oxygen sensing in carotid body glomus cells, aldosterone secretion from adrenal cortex in response to angiotensin II and potassium, sleep, and carcinoma cell proliferation[1–6]. Investigations using genetic manipulation may be limited by compensatory and other changes occurring in knockout animals. For instance, the altered expression pattern of aldosterone synthase in the adrenal gland and compensation by TASK3 in adrenal gland in TASK1 −/− mice cause complexity in understanding specific roles for these channels in aldosterone secretion in adrenal glomerulosa cells [7]. Specific pharmacological modulation of TASK1 over TASK3 (KCNK3 and KCNK9, respectively) could therefore provide important information on the roles of these channels in aldosterone secretion.
There are no published reports of small molecule probes for TASK1 (KCNK3) that possess better than 10-fold selectivity over the closely related TASK3 (KCNK9) channel. The lack of selectivity limits the degree of interrogation one could do to elucidate physiological role(s) of TASK1 in various disease conditions. ML365 is a potent and selective TASK1 inhibitor and thereby a pharmacological tool that can be used to examine the specific roles of TASK1 channels. TASK1 knockout mice displayed impaired T cell proliferation and cytokine production, and markedly reduced axonal degeneration in a multiple sclerosis animal model [8]. ML365 could be a valuable tool for in vivo experiments, and provides a platform for the continued development of improved analogs for in vivo studies of the roles of these channels in autoimmune or CNS diseases. TASK1 is also involved in aldosterone secretion and hypertension [7, 9]. The development of ML365 provides a pharmacological tool to investigate the properties of TASK channels and their specific roles in aldosterone secretion with electrophysiological analysis and biochemical measures of aldosterone. In addition, ML365 may be used to investigate the roles of TASK1 channels in chemosensory control of respiration via central chemosensory neurons and carotid body cells and also in response to hypoxia. Studies utilizing ML365 may help to understand the mechanistic basis of these processes.
2. Materials and Methods
2.1. Assays
The details of the primary HTS and additional assays can be found in the “Assay Description” section in the PubChem BioAssay view under the AIDs as listed (Table 2). Detailed protocols are provided in the Appendix, section 6.3.
Table 2
Summary of assays, listed by title and AID, used for the development of ML365.
2.2. Probe Chemical Characterization
A. Probe Chemical Name, Structure and Physiochemical Data
The IUPAC name of the probe ML365 is 2-methoxy-N-(3-(3-methylbenzamido)phenyl)benzamide. The batch prepared and submitted to the MLSMR is archived as SID 104221970, corresponding to CID 46785920.

B. Structure Verification and Purity: 1H NMR, 13C NMR, LCMS, and HRMS Data
Proton and carbon NMR data for ML365/ SID 104221970/ CID 46785920: Detailed analytical methods and instrumentation are described in Section 2.3, entitled “Probe Preparation” under general experimental and analytical details. The numerical experimental proton and carbon data are represented below, and the experimental proton and carbon spectra are included for reference (Appendix 6, Figures A6A and A6B, respectively).
Proton NMR Data for ML365/ SID 104221970/ CID 46785920:1H NMR (400 MHz, CDCl3) δ 9.86 (s, 1H), 8.25 (dd, J1 = 7.8 Hz, J2 = 1.9 Hz, 1H), 8.14 (br. s, 1H), 8.12 (br. t, J = 2.0 Hz, 1H), 7.67 (br. s, 1H), 7.66 – 7.59 (m, 2H), 7.51 – 7.46 (m, 1H), 7.39 – 7.30 (m, 4H), 7.14 – 7.09 (m, 1H), 7.03 (br. d, J = 8.4 Hz, 1H), 4.05 (s, 3H), 2.59 (s, 3H).
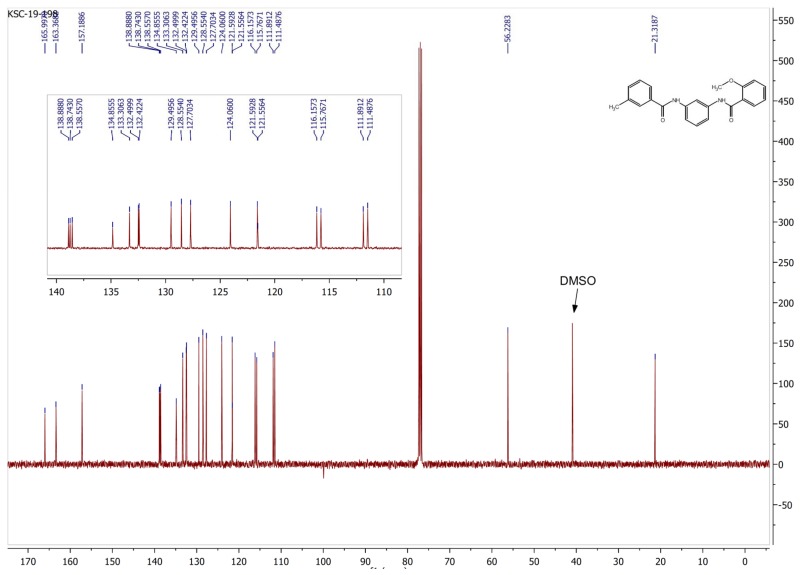
Carbon NMR Data for ML365/ SID 104221970/ CID 46785920:13C NMR (126 MHz, CDCl3) δ 166.00, 163.37, 157.19, 138.89, 138.74, 138.56, 134.86, 133.31, 132.50, 132.42, 129.50, 128.56, 127.70, 124.06, 121.59, 121.56, 116.16, 115.76, 111.89, 111.49, 56.23, 21.32.
LCMS and HRMS Data for ML365/ SID 104221970/ CID 46785920: Detailed analytical methods and instrumentation are described in section 2.3, entitled “Probe Preparation” under general experimental and analytical details. The numerical experimental LCMS and HRMS data are represented as follows: LCMS retention time: 3.159 min. LCMS purity at 214 nm: 100%. HRMS: m/z calcd for C22H20N2O3 (M + H+) 361.1474, found 361.1557. The experimental HRMS and LCMS spectra are included for reference (Appendix, Figure A6C and A6D, respectively).
If available from a vendor, please provide details. This probe is not yet commercially available, as it was designed and prepared as an original analog for this project. A sample of ML365, synthesized at the KU SCC, has been deposited in the MLSMR; however, the compound is also available free of charge from the KU SCC[10].
C. Solubility
Aqueous solubility was measured in phosphate buffered saline (PBS) at room temperature (23°C). PBS by definition is 137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic and a pH of 7.4. Probe ML365 (SID 104221970/ CID 46785920) was found to have a solubility measurement of 0.010 μg/mL, or 0.028 μM, under these conditions. Solubility was also assessed in thallium flux (FDSS) assay medium (1× HBSS pH 7.4), as well as in 140K QPatch assay medium (14 mM NaCl, 140 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 10 mM HEPES, pH 7.4). Under the FDSS assay medium conditions, solubility was determined to be 0.47 μg/mL, or 1.30 μM. Under the QPatch assay medium conditions, solubility was determined to be 0.17 μg/mL, or 0.47 μM[11]. These measurements can be related to the potency values of the assays used in the following ways:
The team recognized the overall modest solubility of ML365; however, the probe criteria (> 10-fold the IC50 value) were met given the potency of the compound. Additional refinements to address this issue will be pursued in the next phase of funding for this project.
D. Stability
Aqueous and Chemical Stability: ML365 was evaluated for stability in PBS, for susceptibility to nucleophilic addition and formation of conjugates when treated with thiol-bearing dithiothreitol (DTT). Figures 3 and 4 represent the time course experiment with ML365 under various conditions. ML365 was dissolved at 10 μM in 1:1 PBS/acetonitrile at pH 7.4 (1% DMSO) and independently incubated at room temperature with no nucleophile present or 50 μM dithiothreitol (DTT). The test reactions were sampled every hour for eight hours (or up to 48 hours, for PBS stability) and analyzed by RP HPLC/UV/HRMS. The analytical RP HPLCUV/HRMS system utilized for the analysis was a Waters Acquity system with UV-detection and mass-detection (Waters LCT Premier). The analytical method conditions included a Waters Acquity BEH C18 column (2.1 × 50mm, 1.8um) and elution with a linear gradient of 99% water to 100% CH3CN at 0.6 mL/min flow rate. Peaks on the 214 nm chromatographs were integrated using the Waters OpenLynx software. Absolute areas under the curve were compared at each time point to determine relative percent compound remaining. All samples were prepared in duplicate and the average plotted[12].

Figure 3
Synthetic scheme for preparation of ML365.
For the aqueous stability experiment, 100% of ML365 was detected remaining after 48 hr in 1:1 PBS/acetonitrile (Figure 1).

Figure 1
Aqueous stability of ML365 over 48 h in 1:1 PBS/acetonitrile.
For the chemical stability experiment, ethacrynic acid, a known Michael acceptor, was used as a positive control and was tested in 1:1 PBS/acetonitrile (Figure 2, panel A). After exposing ML365 to 5-fold concentrations of DTT for 8 h in 1:1 PBS/acetonitrile, 100% of ML365 was remaining (Figure 2, panel B). The masses of potential adducts and dimers of ML365 were searched for in the final samples to determine if any detectable adduct formed or dimerization had occurred. In the case of ML365, no adducts were detected at any time point using LCMS detection[12].

Figure 2
Control experiment with ethacrynic acid (panel A) and chemical stability of ML365 over 8 h in the presence of a 5-fold concentration of dithiothreitol (panel B).
2.3. Probe Preparation
General experimental and analytical details:1H and 13C NMR spectra were recorded on a Bruker AM 400 spectrometer (operating at 400 and 101 MHz respectively) or a Bruker AVIII spectrometer (operating at 500 and 126 MHz respectively) in CDCl3 with 0.03% TMS as an internal standard or DMSO-d6. The chemical shifts (δ) reported are given in parts per million (ppm) and the coupling constants (J) are in Hertz (Hz). The spin multiplicities are reported as s = singlet, bs = broad singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublet and m = multiplet. The LCMS analysis was performed on an Agilent 1200 RRL chromatograph with photodiode array UV detection and an Agilent 6224 TOF mass spectrometer. The chromatographic method utilized the following parameters: a Waters Acquity BEH C-18 2.1 × 50mm, 1.7 um column; UV detection wavelength = 214 nm; flow rate = 0.4ml/min; gradient = 5 – 100% acetonitrile over 3 minutes with a hold of 0.8 minutes at 100% acetonitrile; the aqueous mobile phase contained 0.15% ammonium hydroxide (v/v). The mass spectrometer utilized the following parameters: an Agilent multimode source which simultaneously acquires ESI+/APCI+; a reference mass solution consisting of purine and hexakis(1H, 1H, 3H-tetrafluoropropoxy) phosphazine; and a make-up solvent of 90:10:0.1 MeOH:Water:Formic Acid which was introduced to the LC flow prior to the source to assist ionization. Melting points were determined on a Stanford Research Systems OptiMelt apparatus.
The probe ML365 was synthesized by the method shown (Figure 3). Treatment of nitroaniline 1 with m-toluloyl chloride 2 afforded benzamide 3. Raney nickel catalyzed reduction of 3 afforded the corresponding aniline 4 which was coupled with 2-methoxybenzoyl chloride to deliver the probe ML365 in 13% overall yield. Specific experimental details for the probe are detailed in the section entitled, “Probe Preparation.”
Detailed protocols used for the assembly of ML365 are as follows:

3-methyl-N-(3-nitrophenyl)benzamide. To a solution of 1-amino-3-nitrobenzene (1.0 g, 7.24 mmol) in acetonitrile (10 mL) was added m-toluloyl chloride (1.23 g, 1.05 mL, 7.96 mmol) and triethylamine (0.81 g, 1.11 mL, 7.96 mmol) and the reaction stirred at 70 °C for 3 h. The mixture was then cooled to room temperature, washed with H2O (10 mL), sat NaHCO3 (3 × 10 mL), 1M HCl (3 × 10 mL) and brine (10 mL). The organic layer was dried over MgSO4, filtered, adsorbed to silica and purified by Teledyne ISCO Combiflash chromatography (20 min, 0 – 35% EtOAc:Hex) and the corresponding fractions were collected to produce 3-methyl-N-(3-nitrophenyl)benzamide (1.43 g, 5.58 mmol, 77% yield) as a pale yellow solid. 1H NMR (400 MHz, CDCl3) δ 8.49 (t, J = 2.2 Hz, 1H), 8.16 (br s, 1H), 8.13 (ddd, J1 = 8.2 Hz, J2 = 2.2 Hz, J3 = 1.0 Hz, 1H), 7.99 (ddd, J1 = 8.2 Hz, J2 = 2.2 Hz, J3 = 1.0 Hz, 1H), 7.71 (br s, 1H), 7.69 – 7.64 (m, 1H), 7.54 (t, J = 8.2 Hz, 1H), 7.4 – 7.38 (m, 2H), 2.44 (s, 3H).

N-(3-aminophenyl)-3-methylbenzamide. To a 500 mL round-bottom was added the 3-methyl-N-(3-nitrophenyl)benzamide (0.78 g, 3.06 mmol) with MeOH (10 mL) and CH2Cl2 (10 mL). The reaction was cooled to 0 °C and the Raney nickel (0.018 g, 0.31 mmol) was added. The NaBH4 (0.29 g, 7.64 mmol) was then added portion wise and the reaction stirred at 0 °C for 2.5 h. The reaction was diluted with CH2Cl2 (20 mL) and washed with water (25 mL). The CH2Cl2 layer collected and dried with MgSO4, filtered and adsorbed to silica and purified by Teledyne ISCO Combiflash chromatography (0 – 10% MeOH: CH2Cl2). The corresponding peak was isolated to produce N-(3-aminophenyl)-3-methylbenzamide (0.63 g, 2.77 mmol, 91% yield). 1H NMR (400 MHz, CDCl3) δ 7.74 (br s, 1H), 7.66 (br s, 1H), 7.64 – 7.60 (m, 1H), 7.37 – 7.34 (m, 2H), 7.31 (t, J = 2.1 Hz, 1H), 7.12 (t, J = 8.0 Hz, 1H), 6.79 (ddd, J1 = 8.0 Hz, J2 = 2.0 Hz, J3 = 0.9 Hz, 1H), 6.47 (ddd, J1 = 8.0 Hz, J2 = 2.0 Hz, J3 = 0.9 Hz, 1H), 3.83 (br s, 2H), 2.42 (s, 3H).

2-methoxy-N-(3-(3-methylbenzamido)phenyl)benzamide: ML365, SID 104221970, CID 46785920. To a vial was added the N-(3-aminophenyl)-3-methylbenzamide (0.15 g, 0.67 mmol), acetonitrile (1.5 mL) and triethylamine (0.10 g, 0.14 mL, 1.01 mmol). The 2-methoxybenzoyl chloride (0.14 g, 0.12 ml, 0.81 mmol) was then added and the reaction stirred at 70 °C for 3 h. The reaction was removed from heat and cooled to rt and was quenched with saturated NaHCO3 (5 mL) and extracted with EtOAc (3 × 5 mL). The EtOAc layer was dried with MgSO4, filtered and adsorbed to silica and purified by Teledyne ISCO Combiflash chromatography (30 min, 0 – 35% EtOAc:Hex) and fractions 1 – 5 were collected to produce 2-methoxy-N-(3-(3-methylbenzamido)phenyl)benzamide (0.043 g, 0.119 mmol, 18 % yield) as a waxy solid. 1H NMR (400 MHz, CDCl3) δ 9.86 (s, 1H), 8.25 (dd, J1 = 7.8 Hz, J2 = 1.9 Hz, 1H), 8.14 (br. s, 1H), 8.12 (br. t, J = 2.0 Hz, 1H), 7.67 (br. s, 1H), 7.66 – 7.59 (m, 2H), 7.51 – 7.46 (m, 1H), 7.39 – 7.30 (m, 4H), 7.14 – 7.09 (m, 1H), 7.03 (br. d, J = 8.4 Hz, 1H), 4.05 (s, 3H), 2.59 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 166.00, 163.37, 157.19, 138.89, 138.74, 138.56, 134.86, 133.31, 132.50, 132.42, 129.50, 128.56, 127.70, 124.06, 121.59, 121.56, 116.16, 115.76, 111.89, 111.49, 56.23, 21.32. LCMS Retention time: 3.154 min. LCMS purity 97.4%. HRMS (ESI): m/z calcd for C22H20N2O3 [M+H]+ 361.1474, found 361.1557.
3. Results
The effect of TASK1/KCNK3 inhibitors was titrated in a thallium flux assay (AID 651638, AID 652212) similar to that used in the primary high-throughput screen (AID 602410) and further examined in automated electrophysiology experiments (AID 652209, AID 652210).
3.1. Dose Response Curves for Probe
A. Fluorescent Assay
The primary HTS assay utilized a thallium sensitive dye to measure potassium channel activity in a CHO cell line expressing TASK1/KCNK3 (AID 602410). An extracellular solution containing both thallium and potassium was added triggering thallium influx into the cells. The electrochemical gradient drives the net inflow of thallium down its concentration gradient and the accumulation of intracellular thallium increases the fluorescence of the FluxOR™ dye (Invitrogen). The reported IC50 values of ML365 were determined by averaging multiple independent experiments. Figure 4 illustrates the typical titratable inhibition of TASK1/KCNK3-mediated thallium influx by ML365 in one experiment. After normalization to buffer controls (0% inhibition) and TASK1/KCNK3-independent thallium signal (100% inhibition), the resulting dose-dependent curve was fit with an IC50 value of 4 nM. In replicate experiments (n=4), the mean ± SD of the determined IC50 values were 4.3 ± 0.4 nM.
B. Automated Electrophysiological Assay
To further validate the compound effect on TASK1/KCNK3 channels, an electrophysiological assay was developed on the QPatch 16X automated electrophysiology instrument (Sophion) (AID 652209). Whole cell voltage clamp recordings were made using the same TASK1/KCNK3-expressing CHO cells used in the primary screen. A complete description of the protocol is included in this report within the appendix (Appendix 6.3). Concentration response curves were generated from data collected at −30 mV and normalized to 100% inhibition determined through the application of 2 mM barium in 140 mM potassium solution at pH 5.8. This protocol was used to evaluate select compounds and the data has been collected in the SAR tables. ML365 effect on the TASK1/KCNK3 channel was evaluated in multiple independent experiments using this method. Figure 5A illustrates dose-dependent inhibition of ML365 on whole cell currents in one experiment. Figure 5B summarizes the inhibition of TASK1/KCNK3 currents at −30 mV and shows that ML365 inhibited TASK1/KCNK3 with an IC50 of 12 ± 1 nM (n=4).
3.2. Cellular Activity
The primary HTS assay and all secondary assays are cell-based assays, indicating that ML365 can gain access to its molecular target when applied to cells. The compound did not exhibit acute toxicity in cell-based assays at concentrations up to 30 μM.
In a separate cytotoxicity study, CHO cells were incubated with ML365 for 24 hours at three independent concentrations (1 μM, 3 μM, and 10 μM). Lactate dehydrogenase was measured to estimate the percent of cell death. After 24 hr incubation, ML365 did not cause significant cytotoxicity at concentrations up to 10 μM as compared to control.
3.3. Profiling Assays
A. Profiling Against Host Targets
Probe ML365 was submitted to Eurofins PanLabs (formerly Ricerca Biosciences) to evaluate it in radioligand binding assays against a panel of 68 GPCRs, ion channels and transporters at a single concentration of 10 μM, each in duplicate. The following activities were determined as > 50% inhibition at 10 μM (Table 3). The entire data set is included in the Appendix, section 6.4.
Table 3
Targets that were inhibited by ML365 with > 50 percent inhibition at 10 μM.
B. Channel Selectivity Studies
The selectivity of ML365 for inhibiting the TASK1 potassium channel as compared to block of other related and non-related potassium channels was determined using fluorescent and electrophysiological assays.
C. Selectivity in Fluorescent Experiments
ML365 was evaluated for its ability to block a closely related potassium channel, TASK3 (KCNK9), using HEK293 cell line stably expressing the channel under a tetracycline inducible promoter (AID 652211). TASK3 activity was monitored using a fluorescence assay similar to that described for TASK1. Changes in fluorescence intensity due to modulation of channel activity were recorded on an FDSS instrument (Hamamatsu). Based on this assay, ML365 inhibited TASK3 activity with an IC50 of 0.26 ± 0.02 μM (n=4, Figure 6) The effect of all SAR compounds on a panel of potassium channels was also evaluated using an FDSS instrument (Hamamatsu) measuring changes in fluorescence intensity and the data for all compounds has been collected in the SAR tables including their respective AIDs. ML365 activity against all cell lines tested is summarized in Figure 6 indicating the average percent inhibition of thallium signal in cells treated with 1 μM compound. ML365 showed modest effect against KCNQ2, inhibiting less than 20% of the signal. Little to no inhibition was observed in hERG- and Kir2.1-expressing cells.
D. Selectivity in Electrophysiological Studies
Whole cell voltage clamp recordings were made from HEK293 cells expressing the closely related K2P channel, TASK3, using the QPatch 16X automated electrophysiology instrument (Sophion) (AID 652210). A voltage protocol similar to that used for monitoring TASK1 function was utilized and inhibition of TASK3 current determined at −30 mV. ML365 exhibited modest inhibition of TASK3 with an IC50 40 times that observed for TASK1 (0.012 μM and 0.48 μM, respectively). Figure 7 compares the concentration response curves generated for ML365 against both cell types tested in automated electrophysiology experiments demonstrating the selectivity of ML365 for TASK1 over TASK3.
4. Discussion
4.1. Comparison to existing art and how the new probe is an improvement
Two compounds (A1899 and C23) merit comparison to ML365 on the basis of their TASK1 potency and selectivity over other channels, especially TASK3.
C23:[13] While C23 is reported as a TASK3-selective inhibitor with 10-fold selectivity over TASK1, internal assessment in FDSS and QPatch assays revealed that the C23 was a TASK1-selective inhibitor with 11.2-fold and 2.1-fold selectivity over TASK3 in FDSS and QPatch assays, respectively (Fig. 8). When profiled against hERG, Kir2.1 and KCNQ2 channels, C23 also showed activity on hERG and KCNQ2 at 4.7 and 4.3 μM, respectively. The internally derived TASK1 data from FDSS and QPatch assays showed tremendous congruency (16 and 17 nM, respectively).

Figure 8
Structure and literature and MLPCN determined potency and selectivity data for C23.
A1899:[14] When assessed internally, A1899 demonstrated weaker potencies than those reported, although it did manifest as a TASK1-selective inhibitor and with the same ballpark fold-selectivity over TASK3 in the FDSS assays (Figure 9). A1899 exhibited an FDSS assay TASK1 IC50 value of 0.13 μM and FDSS assay TASK3 IC50 value of 2.02 μM (selectivity index = 15.5-fold for TASK1). In internal QPatch assays, A1899 showed only a 2-fold selectivity for TASK1 over TASK3. No significant liabilities were noted against non-induced parental cells, or hERG, Kir2.1 or KCNQ2 channels.

Figure 9
Structure and literature and MLPCN determined potency and selectivity data for A1899.
ML365 represents a best-in-class probe, as it improves upon both (a) TASK1 potency in both FDSS and QPatch assays, and (b) selectivity over TASK3, as compared to A1899 and C23 with either reported data or when experimentally compared side-by-side against ML365. When compared to C23, ML365 not only possesses a more favorable profile in terms of these parameters, but it also harbors a clean assessment against other channels – a claim one cannot make with C23. Overall, the resulting probe from this effort, ML365, potently inhibits TASK1 to the same degree as that reported for A1899 but with much better selectivity over TASK3 (at least 62-fold), thus advancing capabilities of those working in this area of research to selectively elucidate mechanisms involving TASK1 in a therapeutic context.
5. References
- 1.
- Lauritzen I, et al. K+-dependent cerebellar granule neuron apoptosis. Role of task leak K+ channels. J Biol Chem. 2003;278(34):32068–76. [PubMed: 12783883]
- 2.
- Pei L, et al. Oncogenic potential of TASK3 (Kcnk9) depends on K+ channel function. Proc Natl Acad Sci U S A. 2003;100(13):7803–7. [PMC free article: PMC164668] [PubMed: 12782791]
- 3.
- Bayliss DA, Barrett PQ. Emerging roles for two-pore-domain potassium channels and their potential therapeutic impact. Trends Pharmacol Sci. 2008;29(11):566–75. [PMC free article: PMC2777628] [PubMed: 18823665]
- 4.
- Meuth SG, et al. TWIK-related acid-sensitive K+ channel 1 (TASK1) and TASK3 critically influence T lymphocyte effector functions. J Biol Chem. 2008;283(21):14559–70. [PubMed: 18375952]
- 5.
- Pang DS, et al. An unexpected role for TASK-3 potassium channels in network oscillations with implications for sleep mechanisms and anesthetic action. Proc Natl Acad Sci U S A. 2009;106(41):17546–51. [PMC free article: PMC2751655] [PubMed: 19805135]
- 6.
- Es-Salah-Lamoureux Z, Steele DF, Fedida D. Research into the therapeutic roles of two-pore-domain potassium channels. Trends Pharmacol Sci. 2010;31(12):587–95. [PubMed: 20951446]
- 7.
- Heitzmann D, et al. Invalidation of TASK1 potassium channels disrupts adrenal gland zonation and mineralocorticoid homeostasis. EMBO J. 2008;27(1):179–87. [PMC free article: PMC2206116] [PubMed: 18034154]
- 8.
- Bittner S, et al. TASK1 modulates inflammation and neurodegeneration in autoimmune inflammation of the central nervous system. Brain. 2009;132(Pt 9):2501–16. [PMC free article: PMC3031313] [PubMed: 19570851]
- 9.
- Davies LA, et al. TASK channel deletion in mice causes primary hyperaldosteronism. Proc Natl Acad Sci U S A. 2008;105(6):2203–8. [PMC free article: PMC2538899] [PubMed: 18250325]
- 10.
- Kansas, U.o. Available from: http://www
.scc.ku.edu/ - 11.
- Mangravita-Novo A. U.o. Kansas, editor. 2013.
- 12.
- Porubsky P. U.o. Kansas, editor. 2013.
- 13.
- Coburn CA, et al. Discovery of a pharmacologically active antagonist of the two-pore-domain potassium channel K2P9.1 (TASK-3). ChemMedChem. 2012;7(1):123–33. [PubMed: 21916012]
- 14.
- Streit AK, et al. A specific two-pore domain potassium channel blocker defines the structure of the TASK-1 open pore. J Biol Chem. 2011;286(16):13977–84. [PMC free article: PMC3077598] [PubMed: 21362619]
- 15.
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. [PubMed: 10838414]
- 16.
- Brideau C, et al. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen. 2003;8(6):634–47. [PubMed: 14711389]
6. Appendix
6.1. Quality control 1H-NMR and 13C-NMR spectra for ML365
6.2. Quality control LCMS Purity and Mass ID data: HRMS and HPLC for ML365
6.3. Assay descriptions
HTS for identification of compounds that inhibit the two-pore domain potassium channel TASK1
- 1.
Cell culture: Cells are routinely cultured in Hams F12 medium, supplemented with 10% Fetal Bovine Serum (FBS), 50 IU/ml penicillin, 50 μg/ml streptomycin, 400 μg/ml hygromycin and 10 μg/mL Blasticidin S
- 2.
Cell plating: Add 50 μl/well of 120,000 cells/ml re-suspended in Hams F12 medium with 10% FBS and 1μg/μl Tetracycline
- 4.
Remove medium and add 25 μl/well of 1× FluxOR solution to cells
- 5.
Incubate 90 minutes at room temperature (RT) in the dark
- 6.
Prepare 7.5× compound plates and control plates on Cybi-Well system; test compounds are prepared using assay buffer; controls are assay buffer (EC0), and ECmax of SID17386958
- 7.
Remove FluxOR dye solution and add 20 μl/well of assay buffer to cells
- 8.
Add 4 μl of 7.5× compound stock into the cell plates via Cybi-Well system
- 9.
Incubate all cell plates for 20 minutes at RT in the dark
- 10.
Prepare 5× stimulus buffer containing 25 mM K2SO4 and 7 mM Tl2SO4
- 11.
Load cell plates to Hamamatsu FDSS 6000 kinetic imaging plate reader
- 12.
Measure fluorescence for 10 seconds at 1Hz to establish baseline
- 13.
Add 6 μl/well of stimulus buffer onto cells and continue measuring fluorescence for 180 seconds
- 14.
Calculate ratio readout as F(max−min)/F0
- 15.
Calculate the average and standard deviation for negative and positive controls in each plate, as well as Z and Z′ factors [15]
- 16.
Calculate B scores [16] for test compounds
- 17.
In SAR analysis, IC50 and Hill Constant calculations from replicates were generated using Microcal Origin 6.0
Selectivity assay for the identification of compounds that inhibit the two-pore domain potassium channel TASK1 using TASK3-expressing cells
- Cell culture: Cells are routinely cultured in DMEM/high glucose medium, supplemented with 10% Fetal Bovine Serum (HiFBS), 50 IU/ml penicillin, 50 μg/mL streptomycin, 15 μg/mL Blasticidin S and 400 μg/mL hygromycin
- Cell plating: Add 50 μl/well of 300,000 cells/ml re-suspended in DMEM/high glucose medium with 10% FBS and 1μg/μl Tetracycline
- Incubate overnight at 37°C and 5% CO2
- Remove medium and add 25 μl/well of 1× FluxOR solution to cells
- Incubate 90 minutes at room temperature (RT) in the dark
- Prepare 7.5× compound plates and control plates on Cybi-Well system; test compounds are prepared using assay buffer; controls are assay buffer (EC0), and ECmax of SID17386958
- Remove FluxOR dye solution and add 20 μl/well of assay buffer to cells
- Add 4 μl of 7.5× compound stock into the cell plates via Cybi-Well system
- Incubate all cell plates for 20 minutes at RT in the dark
- Prepare 5× stimulus buffer containing 25 mM K2SO4 and 7 mM Tl2SO4
- Load cell plates to Hamamatsu FDSS 6000 kinetic imaging plate reader
- Measure fluorescence for 10 seconds at 1Hz to establish baseline
- Add 6 μl/well of stimulus buffer onto cells and continue measuring fluorescence for 110 seconds
- Calculate ratio readout as F(max−min)/F0
- Calculate the average and standard deviation for negative and positive controls in each plate, as well as Z and Z′ factors
- IC50 and Hill Constant calculations from replicates were generated using Microcal Origin 6.
Selectivity assay for the identification of compounds that inhibit the two-pore domain potassium channel TASK1 using cells expressing non-K2P potassium channels
- Cell culture: Cells are routinely cultured in DMEM/F12 medium, supplemented with 10% Fetal Bovine Serum (FBS), 50 IU/ml penicillin, 50μg/ml streptomycin, and 500μg/ml G418
- Cell plating: Add 50 μl/well of 300,000 cells/ml re-suspended in DMEM/F12 medium with 10% FBS
- Incubate overnight at 37°C and 5% CO2
- Remove medium and add 25 μl/well of 1× FluxOR solution to cells
- Incubate 90 minutes at room temperature (RT) in the dark
- Prepare 7.5× compound plates and control plates on Cybi-Well system; test compounds are prepared using assay buffer
- Remove FluxOR dye solution and add 20 μl/well of assay buffer to cells
- Add 4 μl of 7.5× compound stock into the cell plates via Cybi-Well system
- Incubate all cell plates for 20 minutes at RT in the dark
- Prepare 5× stimulus buffer containing 25 mM K2SO4 and 7 mM Tl2SO4
- Load cell plates to Hamamatsu FDSS 6000 kinetic imaging plate reader
- Measure fluorescence for 10 seconds at 1Hz to establish baseline
- Add 6 μl/well of stimulus buffer onto cells and continue measuring fluorescence for 110 seconds
- Calculate ratio readout as F(max−min)/F0
- Calculate the average and standard deviation for negative and positive controls in each plate, as well as Z and Z′ factors
- Calculate percent inhibition by normalizing fluorescent ratios to EC0 controls. IC50 and Hill Constant calculations from replicates were generated using Microcal Origin 6.
TASK1 validation assay using automated electrophysiology
- Cell culture: Cells are routinely cultured in F12 medium, supplemented with 10% Fetal Bovine Serum (HiFBS), 10 μg/mL Blasticidin S and 400 μg/mL hygromycin
- The stable CHO cell line expressing inducible TASK1 expression was seeded and induced (1 μg/ml Tetracycline) in 15 cm dish two days before testing on QPatch 16X automated patch clamp system andTASK1 cells were incubated at 30 °C overnight before recording.
- Cells were detached into single cell suspension and allowed to recover for at least 1 hr before running in single-hole mode on QPatch.
- Cells were pipetted to QPlate wells and whole cell recordings established using the instrument’s integrated fluidics and pressure controls and following the manufacturer’s recommended protocols. Whole cell currents were filtered at 1 kHz (4-pole Bessel) and sampled at 5 kHz. The series resistance was compensated by 60% with a cut-off frequency of 0.8 kHz.
- TASK1 cells were in 4 mM potassium external buffer (4K buffer) for 2 minutes after which time external solution was switched to 140 mM potassium external buffer (140K buffer). TASK1 currents were measured by brief voltage ramps applied every 5 seconds. During voltage ramps, cells were held at −80 mV for 50 ms, then at −100 mV for 50 ms followed by a 750 ms ramp to +50 mV, cells were then held at +50 mV for 5 ms, and then back to −80 mV for 40 ms.
- In order to evaluate effects of test compounds as inhibitors of TASK1 channels, cells were stabilized in 4K buffer for 2 minutes. After application of 140K buffer, cells were recorded for 3 minutes. Then test compound in 140K buffer was added to cells in a dual addition mode in which the external solution was completely exchanged twice for each concentration. Compound effects were determined after a 5 minute incubation-recording period. Three test compound concentrations were recorded during each protocol. Compound was washed off by application of 140K buffer for 2 minutes after testing the highest concentration. A reference 140 mM potassium solution containing 2 mM barium at pH 5.8 was then added to cells and recorded for 3 minutes to define the level of 100% inhibition.
- Percent inhibition of TASK1 currents was calculated at the end of each test period as 100 × (1−(Itest-Ibarium/I140k -Ibarium)) where I is the measured current at −30 mV.
- Percent inhibition values calculated at each test compound concentration for a number of cells were combined and fitted with Hill equation to provide IC50 values.
Selectivity assay for the identification of compounds that inhibit the two-pore domain potassium channel TASK1 using automated electrophysiology
- Cell culture: Cells are routinely cultured in DMEM/high glucose medium, supplemented with 10% Fetal Bovine Serum (HiFBS), 15 μg/mL Blasticidin S and 400 μg/mL hygromycin
- The stable HEK 293 cell line expressing inducible TASK3 expression was seeded and induced (0.5μg/ml Tetracycline) in 15 cm dish two days before testing on QPatch 16X automated patch clamp system.
- Cells were detached into single cell suspension and allowed to recover for at least 1 hr before running in single-hole mode on QPatch.
- Cells were pipetted to QPlate wells and whole cell recordings established using the instrument’s integrated fluidics and pressure controls and following the manufacturer’s recommended protocols. Whole cell currents were filtered at 1 kHz (4-pole Bessel) and sampled at 5 kHz. The series resistance was compensated by 20% with a cut-off frequency of 0.2 kHz.
- TASK3 cells were in 4 mM potassium external buffer (4K buffer) for 2 minutes after which time external solution was switched to 140 mM potassium external buffer (140K buffer). TASK3 currents were measured by brief voltage ramps applied every 5 seconds. During voltage ramps, cells were held at −80 mV for 50 ms, then at −100 mV for 50 ms followed by a 600 ms ramp to +20 mV, cells were then held at +20 mV for 5 ms, and then back to −80 mV for 40 ms.
- In order to evaluate effects of test compounds as inhibitors of TASK3 channels, cells were stabilized in 4K buffer for 2 minutes. After application of 140K buffer, cells were recorded for 3 minutes. Then test compound in 140K buffer was added to cells in a dual addition mode in which the external solution was completely exchanged twice for each concentration. Compound effects were determined after a 5 minute incubation-recording period. Three test compound concentrations were recorded during each protocol. Compound was washed off by application of 140K buffer for 2 minutes after testing the highest concentration. A reference 140 mM potassium solution containing 2 mM barium at pH 5.8 was then added to cells and recorded for 3 minutes to define the level of 100% inhibition.
- Percent inhibition of TASK3 currents was calculated at the end of each test period as 100 × (1−(Itest-Ibarium/I140k -Ibarium)) where I is the measured current at −30 mV.
- Percent inhibition values calculated at each test compound concentration for a number of cells were combined and fitted with Hill equation to provide IC50 values.
6.4. Eurofins Profiling Report for ML365
Table 4ML365 profiling data
| Entry | Target | Species | Percent Inhibition |
|---|---|---|---|
| 1 | Adenosine A1 | human | 44 |
| 2 | Adenosine A2A | human | 66 |
| 3 | Adenosine A3 | human | 57 |
| 4 | Adrenergic α1A | rat | 19 |
| 5 | Adrenergic α1B | rat | 9 |
| 6 | Adrenergic α1D | human | 11 |
| 7 | Adrenergic α2A | human | 9 |
| 8 | Adrenergic β1 | human | 4 |
| 9 | Adrenergic β2 | human | 7 |
| 10 | Androgen (Testosterone) | rat | −17 |
| 11 | Bradykinin B1 | human | 0 |
| 12 | Bradykinin B2 | human | −3 |
| 13 | Calcium Channel L-Type, Benzothiazepine | rat | 5 |
| 14 | Calcium Channel L-Type, Dihydropyridine | rat | 6 |
| 15 | Calcium Channel N-Type | rat | −2 |
| 16 | Cannabinoid CB1 | human | 6 |
| 17 | Dopamine D1 | human | −1 |
| 18 | Dopamine D2S | human | 2 |
| 19 | Dopamine D3 | human | 1 |
| 20 | Dopamine D4.2 | human | 71 |
| 21 | Endothelin ETA | human | −2 |
| 22 | Endothelin ETB | human | 0 |
| 23 | Epidermal Growth Factor (EGF) | human | 1 |
| 24 | Estrogen ERα | human | 4 |
| 25 | GABAA, Flunitrazepam, Central | rat | 11 |
| 26 | GABAA, Muscimol, Central | rat | −2 |
| 27 | GABAB1A | human | 9 |
| 28 | Glucocorticoid | human | 19 |
| 29 | Glutamate, Kainate | rat | 0 |
| 30 | Glutamate, NMDA, Agonism | rat | 13 |
| 31 | Glutamate, NMDA, Glycine | rat | −6 |
| 32 | Glutamate, NMDA, Phencyclidine | rat | −5 |
| 33 | Histamine H1 | human | 17 |
| 34 | Histamine H2 | human | 26 |
| 35 | Histamine H3 | human | −1 |
| 36 | Imidazoline I2 | rat | −5 |
| 37 | Interleukin IL-1 | mouse | −1 |
| 38 | Leukotriene, Cysteinyl | human | 2 |
| 39 | Melatonin MT1 | human | 9 |
| 40 | Muscarinic M1 | human | −9 |
| 41 | Muscarinic M2 | human | 7 |
| 42 | Muscarinic M3 | human | −3 |
| 43 | Neuropeptide Y Y1 | human | 0 |
| 44 | Neuropeptide Y Y2 | human | −4 |
| 45 | Nicotinic Acetylcholine | human | 0 |
| 46 | Nicotinic Acetylcholine α1, Bungarotoxin | human | 4 |
| 47 | Opiate δ1 (OP1, DOP) | human | 4 |
| 48 | Opiate κ(OP2, KOP) | human | 69 |
| 49 | Opiate μ(OP3, MOP) | human | 22 |
| 50 | Phorbol Ester | mouse | −18 |
| 51 | Platelet Activating Factor (PAF) | human | −7 |
| 52 | Potassium Channel [KATP] | hamster | 8 |
| 53 | Potassium Channel hERG | human | 24 |
| 54 | Prostanoid EP4 | human | 10 |
| 55 | Purinergic P2X | rabbit | 29 |
| 56 | Purinergic P2Y | rat | 1 |
| 57 | Rolipram | rat | −3 |
| 58 | Serotonin (5-Hydroxytryptamine) 5-HT1A | human | 35 |
| 59 | Serotonin (5-Hydroxytryptamine) 5-HT2B | human | 94 |
| 60 | Serotonin (5-Hydroxytryptamine) 5-HT3 | human | 10 |
| 61 | Sigma σ1 | human | 16 |
| 62 | Sodium Channel, Site 2 | rat | −21 |
| 63 | Tachykinin NK1 | human | 0 |
| 64 | Thyroid Hormone | rat | 5 |
| 65 | Transporter, Dopamine (DAT) | human | 29 |
| 66 | Transporter, GABA | rat | −7 |
| 67 | Transporter, Norepinephrine (NET) | human | 39 |
| 68 | Transporter, Serotonin(5-Hydroxytryptamine) (SERT) | human | −4 |
- a
Present address: UCSF School of Medicine, Department of Physiology, San Francisco, CA
- b
Present address: Department of Biochemistry, Carver College of Medicine, University of Iowa, Iowa City, IA
- c
Present address: Essen BioScience, Ann Arbor, MI
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- ML365: Development of Bis-Amides as Selective Inhibitors of the KCNK3/TASK1 Two ...ML365: Development of Bis-Amides as Selective Inhibitors of the KCNK3/TASK1 Two Pore Potassium Channel - Probe Reports from the NIH Molecular Libraries Program
- Discovery of ML370, an inhibitor of Vibrio cholerae Quorum Sensing Acting via th...Discovery of ML370, an inhibitor of Vibrio cholerae Quorum Sensing Acting via the LuxO response regulator - Probe Reports from the NIH Molecular Libraries Program
- The Role of PHOSPHO1 in the Initiation of Skeletal Calcification - Probe Reports...The Role of PHOSPHO1 in the Initiation of Skeletal Calcification - Probe Reports from the NIH Molecular Libraries Program
- A high throughput screen for inhibitors of nematode detoxification genes - Probe...A high throughput screen for inhibitors of nematode detoxification genes - Probe Reports from the NIH Molecular Libraries Program
- Functional antagonists of the Apelin (APJ) receptor - Probe Reports from the NIH...Functional antagonists of the Apelin (APJ) receptor - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...