NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
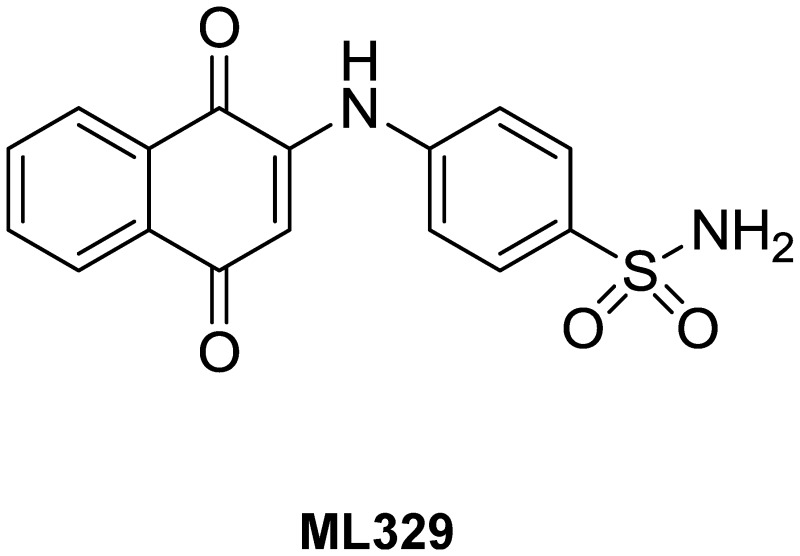
Micropthalmia-associated transcription factor (MITF) is a lineage restricted basic helix-loop-helix leucine zipper transcription factor that is essential for melanocyte development, function and survival. 15% of human melanomas have MITF gene amplification (1). In addition, a vast majority of melanomas are dependent upon MITF for survival. We set out to identify small molecule inhibitors of MITF activity that would allow for better molecular characterization of MITF's role in melanoma. Using an MITF-dependent melanoma cell line, SK-MEL-5, in a cell-based luminescence assay, we measured the promoter activity of a MITF target gene, melastatin (TRPM-1), in a high throughput screen (HTS). 331,578 compounds from the NIH MLPCN compound library were screened. Of these, 3,206 compounds were active (a hit rate of 0.96%). A chloronaphthoquinone (CID 1716436/SID 22416871) was identified in the primary HTS as an inhibitor of TRPM-1 promoter activity. It had potent activity upon retesting in the primary assay, as did several closely related analogs. Structure activity relationship (SAR) studies were performed to improve potency and to minimize deleterious properties. These efforts generated a probe (CID 12387471/ML329) with improved chemical properties and selectivity. In particular, ML329 was not prone to nucleophilic glutathione addition, whereas the initial hit underwent adduct formation. ML329 was tested in two MITF-dependent melanoma cell viability assays, SK-MEL-5 and MALME-3M plus a MITF-independent cell line, A375. ML329 showed specific activity against the MITF-dependent cells, primary melanocytes but no effect on the viability in A375 cells. ML329 reduced the expression of multiple MITF target genes, including pigment-related genes and the cell cycle regulator CDK2. As a tool compound, ML329 will be useful in elucidating the role of MITF in melanocyte lineage development and in melanoma disease progression.
Assigned Assay Grant #: 1 R03 DA031089-01
Screening Center Name & PI: Broad Institute Probe Development Center, Stuart Schreiber
Chemistry Center Name & PI: University of Kansas Specialized Chemistry Center, Jeffrey Aubé
Assay Submitter & Institution: David E. Fisher, Department of Dermatology, Cutaneous Biology Research Center, Massachusetts General Hospital, Harvard Medical School
PubChem Summary Bioassay Identifier (AID): 488944
Probe Structure & Characteristics
| CID/ML# | Target Name | IC50 (μM) [SID, AID] | Anti-target | IC50 (μM) [SID, AID] | Fold Selective | Secondary Assays: IC50 (μM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID 12387471/ML329 | TRPM-1 promoter activity | 1.2 μM [SID 144221520, AID 651588] | A375 cytotoxicity | >35 [SID 144221520, AID 651591] | >30× | 1) SK-MEL-5 cytotoxicity 0.75 μM [SID 144221520, AID 651586] 2) MALME-3M cytotoxicity 0.7 μM [SID 144221520, AID 651585] |
Recommendations for scientific use of the probe
We have developed a small molecule probe (ML329) that inhibits the expression of numerous micropthalmia-associated transcription factor (MITF) target genes and blocks the proliferation of numerous cell lines that require MITF for proliferation. ML329 could directly or indirectly interact with MITF or components of the MITF regulatory network. MITF is a major molecular node in the development, proliferation and maintenance of melanocytes (2). It also has an important role in the progression and persistence of melanomas. As a transcription factor that regulates cell cycle and pigmentation, interference of MITF with ML329 will be useful in characterizing the specific roles of MITF in melanoma and validate blockade of MITF function as a potential treatment of melanoma. The probe will benefit many researchers investigating melanoma and the underlying molecular changes in the early stages of oncogenesis and the subsequent changes in disease progression and metastasis. An inhibitor of MITF will also benefit the study of melanogenesis by parsing out MITF's function in this biological process away from its role in the development of other neural crest derived lineages, like the inner ear and osteoclasts (2,3). MITF has been implicated in clear cell sarcoma and ML329 could be used to determine if it is efficacious in that disease context (4).
1. Introduction
The microphthalmia-associated transcription factor (MITF) was identified as the product of a gene that affects murine coat color. Mice lacking all MITF function are devoid of pigment in fur and in eyes due to a total absence of melanocytes (5). The MITF gene encodes a basic-helix-loop-helix leucine zipper (bHLH-ZIP) transcription factor that homo- or heterodimerizes with the related transcription factors: TFEB, TFE3 and TFEC. These transcription factors, collectively termed the MiT family of transcription factors, are more ubiquitously expressed and unlike MITF are not essential for melanocytic differentiation (6). However, all members of the MiT family bind via their basic domains to identical DNA target sequences containing the canonical E-box promoter element CACGTC or the non-palindromic sequence CACATG.
When its activity is up-regulated in normal melanocytes, MITF initiates a transcriptional program leading to melanocyte differentiation, cell cycle arrest, survival and pigmentation. It directly regulates the transcription of major pigmentation genes, including tyrosinase (7,8), tyrosinase-related protein 1 gene (TYRP-1) (9), dopachrome tautomerase/tyrosinase-related protein-2 (DCT/TYRP-2), QNR-71 (10), silver (11), and AIM1 (12). It has also been suggested that MITF may induce cell cycle arrest during melanocytic differentiation, potentially via transcriptional targeting of the cyclic dependent kinase inhibitors p21, CDKN1A (13) and CDK4A (INK4A) (14). The anti-apoptotic protein Bcl-2 is directly activated by MITF and supports the survival of melanocytes since Bcl-2 knockout results in white coat-color due to melanocyte death (15).
A role for MITF in melanocyte survival is further supported by the consequence of MITF mutation in mice and people: melanocyte death, rather than presence of unpigmented melanocytes. Correspondingly, amplification and over-expression of MITF occurs in 15-20% of melanomas, leading to its designation as a bona fide melanoma oncogene (1). Suppression of MITF activity is lethal to melanomas, and high MITF expression is a poor prognostic factor in melanoma patients, as MITF over-expression is associated with a decrease in 5-year overall survival. Moreover, enforced MITF overexpression was shown to cooperate with the common melanoma oncogene BRAF(V600E) to transform human melanocytes (1). These results indicate that MITF can have either differentiative or tumorigenic effects depending on the cellular context. Whereas physiologic activation of Bcl-2 expression may protect melanocytes (for example, from ultraviolet light), its up-regulation in the context of melanoma may actually contribute to this cancer's notorious chemoresistance.
The above results suggest that small molecule probes that suppress MITF would be useful not only in understanding the biology of context-specific transcriptional control, but also for developing therapeutic strategies for melanoma. With the exception of nuclear hormone receptors, success in directly targeting transcription factors has been very limited (16). Therefore, the identification of upstream druggable pathways that regulate MITF would be important as an alternative therapeutic strategy.
2. Materials and Methods
See subsections for a detailed description of the materials and methods used for each assay.
Materials and Reagents
- Steady-Glo® Luciferase Assay System was purchased from Promega (Catalog No. E2550; Madison, WI)
- CellTiter-Glo® Luminescent Cell Viability Assay was purchased from Promega (Catalog No. G7573; Madison, WI)
- Cells to CT Bulk Lysis reagents purchased from Ambion (Catalog No. 4391851C; Grand Island, NY)
- Cells to CT Bulk RT reagents purchased from Ambion (Catalog No. 4391852C; Grand Island, NY)
- Light Cycler 480 Probes Master purchased from Roche (Catalog No. 4887301001)
- Human GAPD (GAPDH) Endogenous Control VIC/MGB probe/primer limited purchased from Applied Biosystems (Catalog No. 4326317E; Grand Island, NY)
- Human MITF FAM probe/primer set purchased from Applied Biosystems (Catalog No. 4331182 Hs01117294_m1; Grand Island, NY)
- Human TRPM1 probe/primer set purchased from Applied Biosystems (Catalog No. 4331182 Hs00170127_m1; Grand Island, NY)
- Human CDK2 probe/primer set purchased from Applied Biosystems (Catalog No. 4331182 Hs01548894_m1; Grand Island, NY)
- Human DCT probe/primer set purchased from Applied Biosystems (Catalog No. 4331182 Hs01098278_m1; Grand Island, NY)
- Human MLANA probe/primer set purchased from Applied Biosystems (Catalog No. 4331182 Hs00194133_m1; Grand Island, NY)
Cell Lines
The following cell lines were used in this study:
- TRPM-1:luc is a SK-MEL-5 melanoma cell line that expresses firefly luciferase under the control of the melastatin (TRPM-1) promoter. This cell line was used in the primary HTS campaign and generated by the Fisher Lab.
- SK-MEL-5 is the parental cell line to SKMEL5 TRPM-1:luc cells, and does not contain the luciferase reporter. This cell line was obtained from ATCC (Catalog Number HTB-70; Manassas, VA).
- A375 obtained from ATCC (Catalog Number CRL-1619; Manassas, VA) is a melanoma cell line characterized to be independent of MITF for growth and survival
- MALME-3M was obtained from ATCC (Catalog Number HTB-64; Manassas, VA) is a melanoma cell line dependent upon MITF activity for growth and survival.
2.1. Assays
A summary listing of completed assays and corresponding PubChem AID numbers is provided in Appendix A (Table A1). Refer to Appendix B for the detailed assay protocols.
2.1.1. SK-MEL-5 TRPM-1 Luciferase Reporter Assay (Primary Assay AID Nos.: AID 493177, AID 493073, AID 493102, AID 540348, AID 624290, AID 624259, AID 624316, AID 624363, AID 624440, AID 624426, AID 624430, AID 651588, AID 651753)
The TRPM1 luciferase promoter construct was transfected into the SK-MEL-5 melanoma cell line and a stable cell line was generated. This promoter is exquisitely sensitive to MITF over-expression and suppression and contains three canonical E-box motifs within the cloned promoter fragment (17). On day 0, cells were plated at 2,000 cells per well into white, opaque 384 well plates in phenol red-free media. On day 1, cells were treated with compounds or positive control for 24 hours. On day 2, 20 uL of SteadyGlo (Promega) was added per well and luminescence signal was determined with the Perkin-Elmer EnVision plate reader. Primary HTS data were analyzed in Genedata Screener Assay Analyzer. All values were normalized against DMSO treated samples and the positive control (18 μM parthenolide, CID 6473881). For the HTS, the average of two replicates was used to rank order activity and to choose compounds for retests. For dose studies, percent (%) activity was determined for each concentration and the concentration response curves (CRCs) were generated with Genedata Screener's Condoseo.
2.1.2. SK-MEL-5 Cell Cytotoxicity Assay (SA 1: AID Nos.: AID 493240, AID 540347, AID 624289, AID 624315, AID 624366, AID 624427, AID 624429, AID 624428, AID 651586)
SK-MEL-5 cells were treated with compounds for 24 hours, and then cell viability was measured using the CellTiter-Glo Assay (Promega), a luciferase-based reagent that measures cellular ATP levels. The compounds were tested at different concentrations to determine IC50 values. Compounds that were active in the primary assay and toxic below 30 μM at 24 hours were considered for probe development. Data were normalized against DMSO in Genedata Screener's Assay Analyzer. Curves were generated with Genedata Screener's Condoseo and showed percent (%) activity for the individual doses.
2.1.3. A-375 Cell Cytotoxicity Assay (SA1: AID Nos.: AID 540335, AID 540346, AID 624489, AID 624324, AID 624364, AID 624368, AID 624488, AID 624490, AID 624492, AID 651591)
A375 cells were treated with compounds for 24 hours, and then cell viability was measured using the CellTiter-Glo Assay (Promega), a luciferase-based reagent that measures cellular ATP levels. The compounds were tested at different concentrations to determine IC50 values. Compounds that were active in the primary assay and were not toxic below 30 μM at 24 hours were considered for probe development. Data were normalized against DMSO in Genedata Screener's Assay Analyzer. Curves were generated with Genedata Screener's Condoseo and showed percent (%) activity for the individual doses.
2.1.4. MALME-3M Cell Cytotoxicity Assay (SA1: AID Nos.: AID 493191, AID 540339, AID 624299, AID 624362, AID 651584, AID 651585)
MALME-3M cells were treated with compounds for 24 hours, and then cell viability was measured using the CellTiter-Glo Assay (Promega), a luciferase-based reagent that measures cellular ATP levels. The compounds were tested at different concentrations to determine IC50 values. Compounds that were active in the primary assay and toxic below 30 μM at 24 hours were considered for probe development. Data were normalized against DMSO in Genedata Screener's Assay Analyzer. Curves were generated with Genedata Screener's Condoseo and showed percent (%) activity for the individual doses.
2.1.5. qPCR Assay for MITF Expression (SAI: AID 651773)
SK-MEL-5 cells were treated with compounds for 24 hours. Next, cells were lysed with DNase I (Ambion, from Cell to CT Lysis Mix). Lysed cells were delivered to a RT-PCR plate (Ambion, Cells to CT RT Mix) and the plates were then processed for reverse transcription to create cDNA. qPCR was performed by transferring cDNA from the RT-PCR plate to a qPCR plate containing PCR master mix (Roche, Probes Master), FAM Taqman probe/primer set for the target gene (human MITF, Applied Biosystem, 4331182 Hs01117294_m1), VIC Taqman probe/primer set for a house keeping gene (human GAPDH, Applied Biosystems C10228) and water. qPCR plates were cycled using a real-time PCR instrument (Roche, Light Cycler). Using the instrument software, a cycle call was generated when each well enters log phase amplification (Ct). The delta Ct value was determined by subtracting the Ct value of the control gene (GAPDH) from the Ct value of the target gene (MITF) in each well. The delta delta Ct value of each compound treatment was determined by averaging the delta Ct values of the mock well on each plate and subtracting that average from the delta Ct value of each compound well. The compounds were tested at different concentrations to determine IC50 values. Data were normalized against DMSO in Genedata Screener's Assay Analyzer. Curves were generated with Genedata Screener's Condoseo and showed percent (%) activity for the individual doses.
2.1.6. qPCR Assay for TRPM1 Expression (SAI: AID 651770)
Protocol is the same as 2.1.5 except the following primers and probes were used: FAM Taqman probe/primer set for the target gene (human TRPM1, Applied Biosystem, 4331182 Hs00170127_m1), VIC Taqman probe/primer set for a house keeping gene (human GAPDH, Applied Biosystems C10228)
2.1.7. qPCR Assay for CDK2 Expression (SAI: AID 651772)
Protocol is the same as 2.1.5 except the following primers and probes were used: FAM Taqman probe/primer set for the target gene (human CDK2, Applied Biosystem, 4331182 Hs01548894_m1), VIC Taqman probe/primer set for a house keeping gene (human GAPDH, Applied Biosystems C10228)
2.1.8. qPCR Assay for DCT Expression (SAI: AID 651771)
Protocol is the same as 2.1.5 except the following primers and probes were used: FAM Taqman probe/primer set for the target gene (human DCT, Applied Biosystem, 4331182 Hs01098278_m1), VIC Taqman probe/primer set for a house keeping gene (human GAPDH, Applied Biosystems C10228)
2.1.9. qPCR Assay for MLANA Expression (SAI: AID 651795)
Protocol is the same as 2.1.5 except the following primers and probes were used: FAM Taqman probe/primer set for the target gene (human MLANA, Applied Biosystem, 4331182 Hs00194133_m1), VIC Taqman probe/primer set for a house keeping gene (human GAPDH, Applied Biosystems C10228)
2.1.10. Cell proliferation assay of primary human melanocytes (SAI: AID 651920)
Primary human neonatal melanocytes were isolated from discarded foreskins by gentle dispase treatment and grown in Ham's F10 media supplemented with 7% FBS, penicillin/streptomycin/glutamine, 0.1 mM 1-methyl-3-(2-methylpropyl)-7H-purine-2,6-dione (IBMX), 50ng/mL 12-tetradecanoylphorbol 13-acetate (TPA), 1 μM Na3VO4 and 1 μM N(6),2′-O-dibutyryladenosine 3′:5′ cyclic monophosphate (dbcAMP). Cells were plated at 4,000 cells per well of a 384 well plate. On the following day, 10 nL of compound was added per well and incubated for 24 hours. The compounds were tested at different concentrations to determine IC50 values. At the end of compound treatment, cell viability was measured with CellTiter-Glo (Promega) and luminescence measured with the PerkinElmer EnVision plate reader. Data were normalized against DMSO in Genedata Screener's Assay Analyzer. Curves were generated with Genedata Screener's Condoseo and showed percent (%) activity for the individual doses.
2.2. Probe Chemical Characterization
The probe was prepared as described in Section 2.3 and Appendix C. 1H NMR, 13C NMR and HRMS were used to characterize this compound and the results are consistent with the proposed structure. UPLC purity of the material studied was 100% at 214 nm.
Summary of Known Probe Properties in PubChem
| IUPAC Chemical Name | 4-((1,4-dioxo-1,4-dihydronaphthalen-2-yl)amino)benzenesulfonamide |
|---|---|
| PubChem CID | 12387471 |
| Molecular Weight | 328.34 |
| Molecular Formula | C16H12N2O4S |
| XLogP3-AA | 1.4 |
| H-Bond Donor | 2 |
| H-Bond Acceptor | 6 |
| Rotatable Bond Count | 3 |
| Exact Mass | 328.051778 |
| Topological Polar Surface Area | 115 |
2.3. Probe Preparation
The probe (ML329) was synthesized in one step from commercially available 1,4-naphthoquinone and sulfanilamide using cerium(III) chloride heptahydrate as a Lewis acid catalyst as shown in Scheme 1. The reaction was allowed to stir at 75 °C for three days (unoptimized) then dilute citric acid was added to the reaction suspension and the insoluble material was collected by filtration. The filter cake was washed with water, dried, then purified by preparative RPLC.

Scheme 1
Synthesis of the Probe (ML329).
Experimental procedures for the synthesis of the probe can be found in Appendix C.
2.4. Additional Analytical Analysis
ML329 was tested for stability in PBS/1% DMSO (Figure 1A), human serum and mouse serum. ML329 was also tested for solubility and GSH/DTT adduct formation (Figures 1B/1C). All results indicate that ML329 is very stable in PBS/1% DMSO, the GSH/DTT adduct assays, and in the different serums. Experimental procedures for the analytical assays are provided in Appendix D.
3. Results
Probe attributes
- Has an average IC50 value of 1.2 μM (+/- 0.1 μM) in the TRPM-1 reporter assay
- Shows activity in two MITF-dependent melanoma cell lines (SK-MEL-5 & MALME-3M)
- No apparent cytotoxicity in A375, a MITF-independent cell line (EC50= >35 μM).
- Shows dose-dependent inhibition of the expression of multiple MITF target in multiple qPCR assays (TRPM-1, MITF, CDK2, DCT, MLANA)
To identify novel inhibitors of MITF, TRPM-1 promoter activity was measured with an engineered cell line that allows for luminescence detection via the TRPM-1 promoter in a MITF-dependent melanoma cell line, SK-MEL-5. The TRPM-1 promoter responds dynamically to alterations of MITF function and is more sensitive than a standard cell cytotoxicity assay (17). This primary cell-based assay was used in a pilot screen that identified parthenolide, the positive control, as a compound that could decrease the luminescence signal. The assay was optimized for automation and a high throughput screen was pursued. The automated assay was first tested with a validation set of 2,240 compounds. These compounds were tested in duplicate with in-plate neutral (DMSO) and positive controls (18 μM parthenolide). Robustness, reproducibility, and variability parameters were analyzed before initiating the full HTS. The HTS was run over the course of several weeks. Data were normalized relative to controls, and plate patterns were corrected using a multiplicative algorithm in Genedata Screener Assay Analyzer. For each compound, the average of the two replicates was determined and used for subsequent analysis.
Determination of hits required several criteria: only assay plates with a Z′ greater than 0.5 were accepted for analysis, compounds needed to reach 60% inhibition relative to 18 μM parthenolide, and score in fewer than 10% of HTS assays listed in PubChem. In total, 331,578 compounds were screened. Of these, 3,206 compounds were considered active (a hit rate of 0.96%), 613 were inconclusive, and 315,055 were inactive. 1,064 compounds scored in over 10% of assays listed in Pubchem and were excluded from further study. Compounds were clustered based upon chemical structure using a customized script in Pipeline Pilot. Clusters were rated based upon structural liabilities and ranked accordingly. Substructures were analyzed and compared to inactive compounds to identify inactive analogs. Representatives from the more desirable clusters were selected for retest. In addition, a small number of inactive analogs were chosen to provide initial structure/activity relationship (SAR) data during the retest studies. Of these, 1,379 compounds were retested over a range of concentrations to validate activity, and 583 compounds showed dose-dependent inhibition of TRPM-1 expression below IC50 of 10 μM. 261 compounds were below IC50= 5 μM.
Since active compounds produce a decrease in signal in the primary assay, confirmation was required that the reduction in luminescence was not a result of general cytotoxicity by the compound or by inhibition of luciferase itself. Compounds were tested in several melanoma cell lines, including A375, a human melanoma cell line that proliferates despite very low levels of MITF protein (17). In recent studies, siRNA-mediated knockdown of MITF does not alter A375 viability or growth rates and seems to persist independent of MITF regulation (4; Fisher lab, unpublished data). In addition, compounds were tested for cytotoxicity in the parental cell line SK-MEL-5 and another MITF dependent cell line, MALME-3M, to verify dose-dependent inhibition of cell proliferation. Of the compounds that retested in the primary assay, 48 compounds showed the appropriate activity in the SK-MEL-5, MALME-3M and A375 assays.
All available dry powder samples of the remainder qualifying compounds were procured from commercial sources and purity was determined. Only compounds over 90% pure were tested. The compounds were retested in the primary assay and in the cytotoxicity assays. 11 compounds showed an IC50 value of less than 35 μM in A375 cells and were excluded from further consideration. Approximately 44 compounds had an IC50 value below 10 μM in the primary assay. Fifteen chemical scaffolds showed potencies below 10 μM, and three of these were prioritized for follow-up assays. MITF is known to regulate the expression of a number of lineage-specific melanogenesis genes and cell cycle regulators. Therefore, compounds were screened for inhibition of gene expression in qPCR assays for the following genes: MITF, TRPM-1, MLANA, BCL2A, and CDK2. Three scaffolds were put forward as candidates for medicinal chemistry but only one scaffold met all of the assay cutoffs and was amenable to analog production. The lead compound was CID 1716436 (Figure 2).

Figure 2
The structure of MLS000680589 (CID 1716436).
As of 01 November 2012, the original hit (SID 22416871, CID 1716436, MLS000680589, BRD-K45681478-001-06-9) is listed as active in 48 of 617 PubChem assays (6 of which are part of the MITF project). It registered an IC50 value in the primary assay of 4.1 μM (Figure 3A, AID 493177), and showed no cytotoxicity after 24 h in A375 cells (Figure 3B, AID 540335). In addition, it showed potency close to the IC50 cutoff of 10 μM in the SK-MEL-5 and MALME-3M cytotoxicity assays (Figure 3C and 3D, respectively). With its good potency and lack of cytotoxicity in A375 cells, it was prioritized for further development. Analogs were designed and synthesized, eventually leading to the more potent and more soluble probe, ML329 (see Section 3.4).
Table 1Probe and Selected Analogs Assay Performance (Average IC50, μM)
| CID / SID | Broad ID / KU ID | Structure | TRPM-1 promoter assay | A375 CTG | SK-MEL-5 CTG | MALME-3M CTG | TRPM-1 qPCR |
|---|---|---|---|---|---|---|---|
| 12387471 / 144221520 | BRD-K73037408-001-01-5 / KUC111774N-03 |
 | 1.2 | 70 | 0.1 | 0.7 | 0.15 |
| 81124 / 144221523 | BRD-K29842115-001-07-4 / KUC111337N-02 |
 | 5 | 60 | 2.7 | 3.4 | 0.6 |
| 72909 / 144221522 | BRD-K75502546-001-01-9 / KUC111359N-02 |
 | 3.2 | 64 | 8.5 | 7.3 | 2.4 |
| 279009 / 144221524 | BRD-K48101260-001-01-8 / KUC111761N-02 |
 | 6.6 | 70 | 1.2 | 13.5 | IA |
| 56951846 / 144221521 | BRD-K93744531-001-01-2 / KUC111358N-02 |
 | 7.7 | 70 | 14 | 18.6 | 6 |
| 56928031 / 135611193 | BRD-K16604218-001-01-2 / KUC111109N |
 | 41.7 | 64 | 70 | 63 | NT |
NT= not tested, IA=inactive (IC50 >35 μM), Values in bold fail to meet probe criteria
3.1. Summary of Screening Results
Refer to subsections for a detailed description of the results.
In the primary HTS, compounds were active if they decreased TRPM-1 expression as measured by luminescence. The positive control, parthenolide (18 μM), caused a decrease in expression that led to over a 6-fold reduction in signal. Compounds with greater than 60% activity of the positive control were considered actives and chosen for confirmation studies (see Section 3.4 for details). Figure 4 displays the critical path for probe development. To explore SAR, numerous analogs were synthesized and tested. Selected results are shown in Tables 2 to 8 in Section 3.4.

Figure 4
Critical Path for Probe Development.
Table 2
Nascent SAR derived from the HTS results.
Table 8
Round 2 SAR and Hydrogen-substituted Naphthoquinone compounds.
3.2. Dose Response Curves for Probe
Figure 5Dose-dependent Activity of the Probe (ML329)
MLS004556029 (CID 12387471, SID 144221520) was tested across a range of concentrations up to 35 μM in the primary assay and several secondary assays. Concentration response curves were generated with Genedata Screener Condeseo and show normalized percent activity for the individual doses. TRPM-1 promoter assay (AID 651588), IC50= 1.2 μM (A); A375 cytotoxicity assay (AID 651591), IC50>35 μM (B); SK-MEL-5 CellTiter-Glo (AID 651586), IC50= 0.75 μM (C) and MALME-3M CellTiter-Glo (AID 651585), IC50 = 0.88 μM (D). ○=replicate 1, △=replicate 2, ◻=replicate 3, ◇=replicate 4
3.3. Scaffold/Moiety Chemical Liabilities
The solubility of the probe (ML329) was experimentally determined to be 0.4 μM in phosphate buffered saline (PBS) solution (pH 7.4). The probe is stable in PBS buffer solution, with 100% remaining after 48 hours (see Figure 1A). While the probe could be imagined to be reactive toward a thiol nucleophile, the probe was found to be quite stable to glutathione (GSH), with 80% remaining after 6 hours (see Figure 1B), and dithiothreitol (DTT), with 100% remaining after 48 hours (see Figure 1C). The probe is stable in human plasma with approximately 100% remaining after a 5 hour incubation. See the Appendix D for the experimental procedures for the solubility, PBS stability, GSH/DTT stability and plasma stability measurements.
3.4. SAR Tables
The naphthoquinone core was chosen based on potency and selectivity for MITF inhibition, while remaining non-toxic to MITF independent melanocytes. Our focus was on improving the biological activity and chemical stability through analog synthesis. To begin, a more detailed analysis of the primary high throughput (AID 488899) and associated initial screens (AID 493073, AID 493191, AID 493240 and AID 540346) was performed (Table 2). Of the 341,348 compounds reported as tested in AID 488899, there were 347 compounds that contained the benzoquinone substructure without containing the 2-chlorobenzoquinone substructure. This list was further culled by only allowing benzoquinones with a single fusion to another ring (173 compounds after filter). A final structural filter was imposed where structurally complex and/or synthetically challenging compounds were excluded to allow for efficient analog production. The last two filters were based on compound biological promiscuity data: 140/154 compounds had PubChem Active_AID_ratios of ≤ 15%, where the Active_AID_ratio is the ratio of the number of PubChem bioassays where the compound tested active divided by the total number of bioassays in which the compound was tested, as a rough surrogate for general bioactivity promiscuity; 37/140 had a RankScore (a measure of activity) in AID 488899 of at least 60. All of these compounds except one (SID 92764352; CID 386492; RankScore of 66 in AID 488899; not shown in Table 2) had a 1,4-naphthoquinone (or quinoline-5,8-dione) core.
Some of the compounds from the high throughput screen had been tested in the confirmatory and secondary assays and these results are shown in Table 2. Therefore, analysis of the compounds centered around the RankScore and confirmatory activities, where available. Since there was not a strong correlation between these two numbers within the activity ranges of interest (≥60 RankScore and < 15 μM AC50), we used the structures in Table 2 to guide us in forming a small matrix-type library including benzamide, sulfonamide, cyclic amine, and aniline pharmacophores. A selection of these compounds was synthesized and tested, with some additional compounds added to probe the structural requirements and tolerances of the system.
The biological assay data and physical properties of these analogs are presented in Tables 3-8. Characterization data (1H NMR spectra and UPLC chromatograms) of these analogs are provided in Appendix F.
Table 3
Round 1 SAR and Anilino-substituted Naphthoquinone Compounds.
In Table 3, direct chloro substitution on the naphthoquinone core (Table 3, entry 1) gave desirable activity, but these analogs were found to react with glutathione and were removed from further consideration as probe candidates. Maintaining the anilino groups (Table 3, entries 2-11) consistently provided active compounds, although the best anilino compounds were appended with piperidine, piperazine, methoxy or hydrogen substituents (Table 3, entries 2-5). Direct methyl substitution on the naphthoquinone core (Table 3, entry 15) attenuated target potency, but removing the anilino functionality (Table 3, entry 13) maintained desirable potency and selectivity. Appending the naphthoquinone with tertiary nitrogens (Table 3, entry 11) also provided active compounds though potency was lost. Two potential probe candidates were noted from this particular series (i.e., Table 3, entries 4 and 13), after the first round SAR feedback.
Table 4 illustrates that replacing the anilino moiety with N-methylacetamides provided an increase in potency, but at the expense of increasing toxicity shown by the antitarget activity (Table 4, entries 1-6). N-methylacetamides were removed from further consideration due to their associated toxicity.
Table 4
Round 1 SAR and N-methylacetamide-subsituted Naphthoquinone Compounds.
Next, we evaluated a series of benzoyl substitutions on the naphthoquinone ring, and the results are summarized in Table 5. Direct comparison of anilino compounds (e.g., Table 3, entries 2, 6, and 9) with their benzoyl counterparts (e.g., Table 5, entries 1, 5, and 6) illustrates that the benzoyl series gave comparable target potencies, while the antitarget potencies often diminished below the advantageous absolute cut-off limits (e.g., Table 5, entries 1-3 and 5). Appending the benzoyl moiety with electron-withdrawing groups resulted in similar activity (Table 5, entry 2) as did extending the alkyl chain pendant to the piperazine ring (Table 5, entry 3). However, a phenyl substitution on the piperazine ring led to a significant loss in potency (Table 5, entry 6; Table 3, entry 9).
Table 5
Round 1 SAR and Benzoyl-substituted Naphthoquinone compounds.
Table 6 contains data from benzene- and thiophene-substituted sulfonamide analogs. Halogen substitutions improved the activities of thiophene analogs (Table 6, entry 3 and 4) but were detrimental to other sulfonamides (Table 6, entry 1 and 2). Naphthoquinones with -H, -OMe, and –NH2 substitutions proved equipotent to the piperazine analogs (Table 6, entries 5, 6, 8, and 10) but piperidine substitution attenuated potency (Table 6, entry 7).
Table 6
Round 1 SAR Benzene- and Thiophene-substituted Naphthoquinone Compounds.
Appending electron-withdrawing groups to the anilino moiety, as shown in Table 7, gave a slight increase in potency (Table 7, entries 1 and 2) compared to a slight loss of activity when electron-donating groups were used (Table 7, entries 1 and 5). Potencies were improved further by exchanging the N-methylpiperazines for N-ethyl- or N-isopropylpiperazines (e.g., Table 7, entries 5-7). The piperidine and pyrrolidine substituents were generally more potent towards the antitarget than the piperazines (Table 7, entries 8-11) and the morpholino-like analogs lost target potency when compared in the SKMEL5 and MALME-3M assays (Table 7, entries 12 and 13). Anilino analogs presented in Table 7 had enhanced potencies over those presented in Table 3, but still did not satisfy the probe criteria.
Table 7
Round 2 SAR and Anilino- and Nitrogen-heterocycle-substituted Naphthoquinone Compounds.
Table 8 shows the SAR results of the hydrogen-substituted naphthoquinones, which were designed and synthesized based on the two potential probe candidates from the first round SAR (Table 8, entries 1 and 3). Adding heteroatoms into the naphthoquinone ring potentiated the antitarget potency (Table 8, entry 4). Ortho- and para-electron-withdrawing groups pendant to the anilino moiety were well tolerated (Table 8, entries 6, 9, and 11) but meta- groups attenuated potencies (Table 8, entries 7 and 10). para-Chloro substitution also attenuated target potency (Table 8, entry 8). Extending the aromatic ring with a methylene linker was a significant detriment to activity (compare Table 8, entries 8 and 12). Substitution at the para-position with a sulfonamide resulted in the most potent compound that retained selectivity and therefore, was nominated as the probe (Table 8, entry 9).
3.5. Cellular Activity
The primary assay and all of the secondary assays were cell-based, all lead compounds showed activity in cells. In the medicinal chemistry phase of our studies, ML329 and its analogs were tested in multiple cell-based experiments. ML329 consistently showed low micromolar activity in multiple cell-based assays and has good permeability and solubility properties. ML329 also showed potent activity in primary human melanocytes (IC50= 7 μM, AID 651920) (Appendix I). The counterscreen with A375 cells helped to remove compounds that possessed non-specific cellular toxicity.
3.6. Profiling Assays
ML329 was run in a panel of assays with PanLabs/Eurofins and showed little to no inhibition with the exception of human Adenosine A2A where it showed 50% inhibition at 10 μM. See Appendix H for more details.
4. Discussion

Figure 6A visual summary of SAR performed for ML329
The aim of this project was to identify suppressors of MITF activity via a robust and sensitive primary screen. We identified ML329, a potent probe that inhibits the growth of multiple MITF-dependent cell lines, reduces the expression of MITF and several MITF-regulated genes. ML329 meets all of the probe attributes and these are listed in Table 9. The long-term goal is to use these probes in elucidating the molecular pathways regulating MITF activity with the goal of translating this knowledge into therapeutic opportunities.
Table 9
Comparison of the Probe to Project Criteria.
Approximately 160 compound analogs were synthesized during this probe development project, of which all are reported in PubChem, and only a subset are reported in this report. ML329 is not part of the current MLSMR HTS library and thus not listed in any PubChem assays. A number of closely related analogs are present in PubChem with various levels of activity. The initial hit (MLS000680589/CID 1716436) has been tested in approximately 617 assays and is active in 48 assays. Six of those assays are part of the MITF project. MLS000680589 was retested in a number of assays with reported activity of less than 10 μM including: a delayed death malaria plastid assay (AID 504832), a qHTS assay for inhibitors of fructose-1,6-bisphosphate aldolase in Giardia lamblia (AID 2451), a Sentrin-specific protease Caspase-6 selectivity assay (AID 488901), an inhibitor in a DnaB-Intein splicing assay (AID 449749), activator of integrin-mediated alleviation of muscular dystrophy (AID 624291), inhibitor of MBNL-1-poly(CUG) RNA binding (AID2675), lipid storage modulation in Drosophila S3 cells (AID 2685), inhibition of beta cell apoptosis (AID 449756) and as an activator of Nrf2 (AID 624171). Many of these are “loss of signal” luciferase assays which could raise concern that there is modulation of luciferase but activity in some of these assays leads to an increase in luciferase signal, like the beta cell apoptosis project (AID 449756), rather than decreased signal as seen in other assays. These results suggest that our lead does not specifically alter luciferase signal but is working by some other means. To address off-target effects, ML329 was tested against a panel of 67 targets by Eurofins Panlabs. The only appreciable inhibition observed was against several human adenosine receptors (Appendix H). The probe compound has potent activity against several melanoma cell lines (e.g. SK-MEL-5, MALME-3M) and primary melanocytes all of which require MITF for survival. The Fisher lab has additional melanoma cell lines to test which will provide further correlative evidence linking compound activity to MITF dependency.
4.1. Comparison to existing art and how the new probe is an improvement
Investigation into relevant prior art entailed searching the following databases: SciFinder and the Thomson Reuters Integrity. The search terms applied and hit statistics for the prior art search are provided in Table A2 (Appendix G). Abstracts were obtained for all references returned and were analyzed for relevance to the current project. For all references that were deemed relevant, the articles were analyzed and the results are summarized below. The searches are current as of October 9, 2012.
Table A2
Search Strings and Databases Employed in the Prior Art Search.
Since the start of the project, a modest amount of small-molecule prior art has been reported in the scientific literature, especially in the year 2012. However, it is not known if these small-molecule MITF inhibitors recapitulate genetic manipulation or deletion of MITF. Prior to the start of this probe project, the pilot screen identified a compound, parthenolide, that decreased TRPM-1 expression but was inadequate in the other cell-based assays because of a lack of killing in MITF-dependent cell lines. It was used as the positive control for the primary assay for this project. This compound does not show selectivity to MITF and potently kills some MITF-independent melanoma and other cancer cell lines. 2-amino-3H-phenoxazin-3-one (APO) was shown to reduce melanogenesis and the expression of MITF (18). We tested APO and found that it potentially inhibited TRPM-1 promoter activity (IC50= 2 μM) but also killed A375 cells (IC50= 9 μM) suggesting a mode of action nonspecific to MITF. Another report described “compound-17”, a small molecule that inhibited MITF binding to E-box DNA motifs that are found in the promoters of several pigment cell-specific genes like TRPM-1 (19). Compound-17 showed activity with 1 mM of compound in an electrophoretic mobility shift assay (EMSA) but did not alter TRPM-1 protein levels with up to 50 μM of compound after 72 hours of treatment. Although we have not measured TRPM-1 protein levels, we have measured promoter activity in the luciferase assay and mRNA transcript level by qPCR. We detect changes with low micromolar amounts of ML329 after 24 hours of treatment in both the luciferase and qPCR TRPM-1 assays (Figure 5A and 8A). One lead compound (CID 9616928) was eliminated because of lack of potency in SK-MEL-5 cell cytotoxicity, but notably, is similar to the HDAC inhibitor, panobinostat/LBH589 (CID 6918837), which has been shown to reduce MITF expression levels and reduce melanoma tumor growth in vivo (20). LBH589 potently reduced TRPM-1 expression in the luciferase reporter assay but failed to meet the other probe criteria for the remaining cell-based assays. In particular, LBH589 was not potent enough in SK-MEL-5 cells and led to over 50% reduced viability in A375 cells. A number of compounds have been reported to inhibit melanogenesis or MITF but none have been shown to directly interact with MITF and so were not tested against ML329. Several of these substances with low micromolar activity are listed in Table 10.
Table 10
Compounds reported to inhibit MITF.
4.2. Mechanism of Action Studies
The assays utilized in this project were phenotypic, cell-based assays that leave some ambiguity to how ML329 works in cells. The assays used in this project only show circumstantial evidence of ML329 regulating MITF and its molecular pathways. The compound could be acting on an upstream activator of MITF since we see a reduction of MITF transcription and several of its target genes by qPCR (Figure 8B, AID 651773). MITF expression is regulated by a number of transcriptional regulators including: MITF itself, BRAF, WNT signaling, SOX10, CREB, and Pax3 (2). ML329 reduced the expression of multiple MITF target genes suggesting a mechanism that impacts a broader profile of MITF targets, including both melanogenesis and cell cycle genes such as CDK2 (Figure 8C). ML329 consistently lowered the expression of several MITF target genes including: melastatin (TRPM-1), dopachrome tautomerase/tyrosinase-related protein-2 (DCT), the cell cycle regulator cyclin-dependent kinase-2 (CDK2) and melan-A (MLANA/MART1). The primary screen was carried out in a BRAF(V600E) mutated melanoma cell line, SK-MEL-5. It is quite clear that ML329's activities are very different from those of BRAF inhibitors, something studied in the Fisher lab quite extensively. BRAF-MEK-MAPK lead to proteolysis of MITF, and thus MAPK pathway suppression leads to MITF stabilization/up-regulation. We have observed (and published) that multiple MITF target genes are up-regulated following BRAF inhibitor treatments. In contrast we find that ML329 suppresses both MITF and multiple of its transcriptional target genes. We believe that MITF up-regulation following BRAF suppression represents a survival mechanism that limits efficacy of BRAF targeted therapies. Therefore, it is plausible that concurrent use of MITF antagonists (like ML329) may offer significant benefit in combination with BRAF inhibitors. Proper determination of mechanism of action will require a number of different studies, some of which are outlined in Section 4.3.
4.3. Planned Future Studies
Our recommendations for future SAR expansion around the probe (ML329) involve the following: (1) substitute the anilino nitrogen with various alkyl or acyl groups, (2) append the sulfonamide nitrogen with mono- and bis-substituted amines (i.e., methyl, morpholine, piperidine, or pyrrolidine), (3) move the sulfonamide around the ring to which it is currently attached, (4) add substituents around the ring system adjacent to the sulfonamide, (5) replace the aniline-benzenoid moiety with other heterocyclic rings, (6) substitute the C3-position of the naphthoquinone ring system with methyl and methoxy, (7) replace the benzenoid ring of the naphthoquinone core with various heterocyclic rings (i.e., pyridine and piperidine), and (8) substitute the benzenoid ring of the naphthoquinone core with mono- and multi- substituted electron-withdrawing and/or electron-donating groups, as well as groups to probe steric requirements.
ML329 was tested with three melanoma cell lines, primary melanocytes, the TRPM-1 reporter cell line and A375 cells. Only one of these cell lines, A375, does not rely upon MITF for proliferation. There are a number of other melanoma cell lines available (e.g. M14, UACC62, UACC257, 501mel, SK-MEL-28, & SK-MEL-2) and the probe will be tested in these other melanoma cell lines. In addition, MITF has been implicated in clear cell sarcoma and ML329 will be tested in tumor-derived sarcoma cell lines to determine if it maintains activity in a different disease context (4). ML329 was tested with primary melanocytes from a single human donor and will be tested with other donor-derived melanocytes to verify consistency across several genetic backgrounds.
For mechanism of action studies, ML329 could be tested using biophysical experiments to determine direct binding of the compound to the MITF protein. Differential scanning fluorimetry (DSF) or thermal shift is a means of detecting binding of a ligand to purified protein. This technique is routinely used at the Broad Institute and could be applied to MITF. ML329 leads to a decrease in the expression of MITF and a number of target genes. It is not known if the DNA binding capacity of MITF is impaired. ML329 could prevent binding of MITF to E-box domains within promoter elements. An EMSA experiment could address whether binding of MITF to E-box DNA sequences are impacted by the presence of the compound. A weak MITF inhibitor has reported activity in an MITF/E-box EMSA (19). A recurrent mutation in MITF has been associated with familial and sporadic melanoma. This mutation correlates with impaired sumoylation (20). We can test the compound in the context of this mutation to determine if the potency is altered or if inhibition is maintained with this mutant version of MITF.
A gene-expression profiling experiment of SK-MEL-5 cells in the absence or presence of the ML329 will be performed. We plan to test ML329 in a Luminex-based platform called L1000 performed at the Broad Institute where gene expression of 1000 genes is measured after 6 and 24 hours of compound treatment in a panel of 20 cell lines as part of the LINCS program (https://commonfund.nih.gov/LINCS/). Tools developed at the Broad Institute, such as GenePattern and Gene Set Enrichment Analysis (GSEA), will be applied to determine the comparative markers responsible for the greatest difference between the different states. A computational method also developed at the Broad called the Connectivity Map, or cmap, that facilitates target identification through the analysis of small-molecule signatures will also be applied (43). This tool uses gene-expression profiles in a systematic approach to enable the discovery of functional connections between diseases, genetic perturbations, and small-molecule perturbations.
A number of melanoma models that utilize xenografts on rodents are used to determine the capacity of a drug or probe on melanoma (44). These studies can be done by topical administration of the test substance onto the tumor or intraperitoneal injection. We can evaluate ML329 in an analogous tumor graft model.
5. References
- 1.
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. [PubMed: 16001072]
- 2.
- Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. [PubMed: 10898786]
- 3.
- Weilbaecher KN, Motyckova G, Huber WE, Takemoto CE, Hemesath TJ, Xu Y, Hershey CL, Dowland NR, Wells AG, Fisher DE. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitfmi/mi mice. Molecular Cell. 2001;8(4):749–58. [PubMed: 11684011]
- 4.
- Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, Linehan WM, Kung AL, Fisher DE. Oncogenic MITF dysregulation in clear cell sarcoma: Defining the MiT family of human cancers. Cancer Cell. 2006;9(6):473–484. [PubMed: 16766266]
- 5.
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes & Development. 1994;8:2770–2780. [PubMed: 7958932]
- 6.
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. [PubMed: 15568981]
- 7.
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia- associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1995;15:1833. [PMC free article: PMC230408] [PubMed: 7862173]
- 8.
- Yasumoto K, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem. 1997;272:503–509. [PubMed: 8995290]
- 9.
- Fang D, Tsuji Y, Setaluri V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002;30:3096–3106. [PMC free article: PMC135745] [PubMed: 12136092]
- 10.
- Turque N, Denhez F, Martin P, Planque N, Bailly M, Begue A, Stehelin D, Saule S. Characterization of a new melanocyte-specific gene (QNR-71) expressed in v-myc- transformed quail neuroretina. EMBO J. 1996;15:3338–3350. [PMC free article: PMC451897] [PubMed: 8670835]
- 11.
- Du J, Miller AJ, Widlund HR, Horstmann MA, Ramaswamy S, Fisher DE. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. The American Journal of Pathology. 2003;163:333–343. [PMC free article: PMC1868174] [PubMed: 12819038]
- 12.
- Du J, Fisher DE. Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J Biol Chem. 2002;277:402–406. [PubMed: 11700328]
- 13.
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. [PubMed: 15716956]
- 14.
- Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol. 2005;168:35–40. [PMC free article: PMC2171666] [PubMed: 15623583]
- 15.
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, Jordan SA, Jackson IJ, Korsmeyer SJ, Golub TR, Fisher DE. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. [PubMed: 12086670]
- 16.
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. [PMC free article: PMC2951323] [PubMed: 19907488]
- 17.
- Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Research. 2004;64:509–516. [PubMed: 14744763]
- 18.
- Miyake M, Yamamoto S, Sano O, Fujii M, Kohno K, Ushio S, Iwaki K, Fukuda S. Inhibitory effects of 2-amino-3H-phenoxazin-3-one on the melanogenesis of murine B16 melanoma cell line. Biosci. Biotechnol. Biochem. 2010;74(4):753–758. [PubMed: 20445320]
- 19.
- Um JM, Kim HJ, Lee Y, Choi CH, Hoang Nguyen D, Lee HB, Shin JH, Tai No K, Kim EK. A small molecule inhibitor of Mitf-E-box DNA binding and its depigmenting effect in melan-a cells. J Eur Acad Dermatol Venereol. 2012 Oct;26(10):1291–7. [PubMed: 21957942]
- 20.
- Yokoyama S, Feige E, Poling LL, Levy C, Widlund HR, Khaled M, Kung AL, Fisher DE. Pharmacologic suppression of MITF expression via HDAC inhibitors in the melanocyte lineage. Pigment Cell Melanoma Res. 2008 Aug;21(4):457–63. [PMC free article: PMC3771662] [PubMed: 18627530]
- 21.
- Villareal MO, Han J, Yamada P, Shigemori H, Isoda H. Hirseins inhibit melanogenesis by regulating the gene expression of the MITF and melanogenesis enzymes. Exp. Dermatol. 2009;19:450–457. [PubMed: 19765058]
- 22.
- Villareal MO, Han J, Ikuta K, Isoda H. Mechanism of MITF inhibition and morphological differentiation effects of hirsein A on B16 melanoma cells revealed by DNA microarray. J. Dermatol. Sci. 2012;67:26–36. [PubMed: 22564683]
- 23.
- Li X, Guo Y, Sun Y, Zhou J, Gu Y, Li Y. Baicalein inhibits melanogenesis through activation of the ERK signaling pathway. Int. J. Mol. Med. 2010;25:923–927. [PubMed: 20428797]
- 24.
- Huh S, Jung E, Lee J, Roh K, Kim JD, Lee J, Park D. Mechanism of melanogenesis inhibition by propafenone. Arch. Dermatol. Res. 2010;302:561–565. [PubMed: 20549222]
- 25.
- Oh EY, Jang JY, Choi YH, Choi YW, Choi BT. Inhibitory effects of 1-O-methyl-fructofuranose from Schisandra chinensis fruit on melanogenesis in B16F0 melanoma cells. J. Ethnopharmacol. 2010;132:219–224. [PubMed: 20723590]
- 26.
- Chou TH, Ding HY, Lin RJ, Ling JY, Liang CH. Inhibition of melanogenesis and oxidation by protocatechuic acid from Origanum vulgare (Oregano). J. Nat. Prod. 2010;73:1767–1774. [PubMed: 20973550]
- 27.
- Bolton T, Puissant A, Cheli Y, Tomic T, Giuliano S, Fajas L, Deckert N, Ortonne JP, Bertolotto C, Tartare-Deckert S, Ballotti R, Rocchi S. Ciglitazone negatively regulates CXCL1 signaling through MITF to suppress melanoma growth. Cell Death and Differentiation. 2011;18:109–121. [PMC free article: PMC3131866] [PubMed: 20596077]
- 28.
- Syed DN, Afaq F, Maddodi N, Johnson JJ, Sarfaraz S, Ahmad A, Setaluri V, Mukhtar H. Inhibition of human melanoma cell growth by the dietary flavonoid fisetin is associated with disruption of Wnt/β-catenin signaling and decreased MITF levels. J. Invest. Dermatol. 2011;131:1291–1299. [PMC free article: PMC3166244] [PubMed: 21346776]
- 29.
- Lee J, Cho B, Jun Hj, Seo WD, Kim DW, Cho KJ, Lee SJ. Momilactione B inhibits protein kinase A signaling and reduces tyrosinase-related proteins 1 and 2 expression in melanocytes. Biotechnol. Lett. 2012;34(5):805–812. [PubMed: 22215377]
- 30.
- Kim EGE, Min Ji. KR2012016847. Microphthalmia transcription factor inhibitor used in skin-whitening cosmetic composition. 2012
- 31.
- Jin ML, Park SY, Kim YH, Park G, Son HJ, Lee SJ. Suppression of α-MSH and IBMX-induced melanogenesis by cordycepin via inhibition of CREB and MITF, and activation of PI3K/Akt and ERK-dependent mechanisms. Int. J. Mol. Med. 2012;29(1):119–124. [PubMed: 21972008]
- 32.
- Park S, Jin M, Kim Y, Kim Y, Lee SJ. Aromatic-turmerone inhibits α-MSH and IBMX-induced melanogenesis by inactivating CREB and MITF signaling pathways. Archives of Dermatological Research. 2011;303(10):737–744. [PubMed: 21660443]
- 33.
- Kim DS, Lee HK, Park SH, Chae CH, Park KC. AVS-1357 inhibits melanogenesis via prolonged ERK activation. Die Pharmazie. 2009;64(8):532–7. [PubMed: 19746843]
- 34.
- Kim JH, Baek SH, Kim DH, Choi TY, Yoon TJ, Hwang JS, Kim MR, Kwon HJ, Lee CH. Downregulation of melanin synthesis by haginin A and its application to in vivo lightening model. J. Invest. Dermatol. 2008;128(5):1227–35. [PubMed: 18037902]
- 35.
- Huang YH, Lee TH, Chan KJ, Hsu FL, Wu YC, Lee MH. Anemonin is a natural bioactive compound that can regulate tyrosinase-related proteins and mRNA in human melanocytes. J. Dermatol Sci. 2008;49(2):115–23. [PubMed: 17766092]
- 36.
- Joung HS, Song KH, Kim AK. Antimelanogenic effect of taurine in murine melanoma B16F10 cells. Yakhak Hoechi. 2007;51(5):350–354.
- 37.
- Lee HE, Kim EH, Choi HR, Sohn UD, Yun HY, Baek KJ, Kwon NS, Park KC, Kim DS. Dipeptides Inhibit Melanin Synthesis in Mel-Ab Cells through Down-Regulation of Tyrosinase. The Korean Journal of Physiology & Pharmacology: official journal of the Korean Physiological Society and the Korean Society of Pharmacology. 2012;16(4):287–91. [PMC free article: PMC3419765] [PubMed: 22915995]
- 38.
- Kim YJ, No JK, Lee JS, Kim MS, Chung HY. Antimelanogenic activity of 3,4-dihydroxyacetophenone: inhibition of tyrosinase and MITF. Biosci. Biotechnol. Biochem. 2006;70(2):532–4. [PubMed: 16495675]
- 39.
- Park SH, Kim DS, Kim WG, Ryoo IJ, Lee DH, Huh CH, Youn SW, Yoo ID, Park KC. Terrein: a new melanogenesis inhibitor and its mechanism. Cell Mol. Life Sci. 2004;61(22):2878–85. [PubMed: 15558216]
- 40.
- Kim DS, Jeong YM, Park IK, Hahn HG, Lee HK, Kwon SB, Jeong JH, Yang SJ, Sohn UD, Park KC. A new 2-imino-1,3-thiazoline derivative, KHG22394, inhibits melanin synthesis in mouse B16 melanoma cells. Biol. Pharmaceut. Bull. 2007;30(1):180–3. [PubMed: 17202683]
- 41.
- Cho M. Cardamonin suppresses melanogenesis by inhibition of Wnt/beta-catenin signaling. Biochem. Biophys. Res. Comm. 2009;390(3):500–5. [PubMed: 19800318]
- 42.
- Kikuchi T, Zhang J, Huang Y, Watanabe K, Ishii K, Yamamoto A, Fukatsu M, Tanaka R, Akihisa T. Glycosidic Inhibitors of Melanogenesis from Leaves of Momordica charantia. Chem. Biodivers. 2012;9(7):1221–1230. [PubMed: 22782871]
- 43.
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35. [PubMed: 17008526]
- 44.
- Feige E, Yokoyama S, Levy C, Khaled M, Igras V, Lin RJ, Lee S, Widlund HR, Granter SR, Kung AL, Fisher DE. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc Natl Acad Sci U S A. 2011;108(43):E924–33. [PMC free article: PMC3203758] [PubMed: 21949374]
Appendix A. Assay Summary Table
Table A1Summary of Completed Assays and AIDs
| PubChem AID No. | Type | Target | Concentration Range (μM) | Samples Tested |
|---|---|---|---|---|
| 488944 | Summary | MITF Inhibitor project | NA | NA |
| 488899 | Cell-based | TRPM-1 promoter activity assay | 12.5 | 331,578 |
| 493177 | Cell-based | TRPM-1 promoter activity assay | 0.015-35 | 1,241 |
| 493073 | Cell-based | TRPM-1 promoter activity assay | 0.015-35 | 1,241 |
| 493102 | Cell-based | TRPM-1 promoter activity assay | 0.015-35 | 1,241 |
| 493240 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.015-35 | 1,280 |
| 540335 | Cell-based | A375 cytotoxicity assay | 0.015-35 | 1,280 |
| 493191 | Cell-based | MALME-3M cytotoxicity assay | 0.015-35 | 1,280 |
| 540348 | Cell-based | TRPM-1 promoter activity assay | 0.015-35 | 29 |
| 624290 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 70 |
| 624259 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 70 |
| 624316 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 107 |
| 624363 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 57 |
| 624440 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 26 |
| 624426 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 26 |
| 624430 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 62 |
| 651588 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 37 |
| 651753 | Cell-based | TRPM-1 promoter activity assay | 0.00006-35 | 20 |
| 540347 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.015-35 | 30 |
| 624289 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 107 |
| 624315 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 107 |
| 624366 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 57 |
| 624427 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 26 |
| 624429 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 26 |
| 624428 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 62 |
| 651586 | Cell-based | SK-MEL-5 cytotoxicity assay | 0.00006-35 | 37 |
| 540346 | Cell-based | A375 cytotoxicity assay | 0.015-35 | 1,280 |
| 624489 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 107 |
| 624324 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 107 |
| 624364 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 120 |
| 624368 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 57 |
| 624488 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 26 |
| 624490 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 26 |
| 624492 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 62 |
| 651591 | Cell-based | A375 cytotoxicity assay | 0.00006-35 | 37 |
| 540339 | Cell-based | MALME-3M cytotoxicity assay | 0.015-35 | 30 |
| 624299 | Cell-based | MALME-3M cytotoxicity assay | 0.00006-35 | 107 |
| 624362 | Cell-based | MALME-3M cytotoxicity assay | 0.00006-35 | 16 |
| 651584 | Cell-based | MALME-3M cytotoxicity assay | 0.00006-35 | 26 |
| 651585 | Cell-based | MALME-3M cytotoxicity assay | 0.00006-35 | 37 |
| 651773 | Cell-based | SK-MEL-5 qPCR for MITF | 0.00006-35 | 35 |
| 651770 | Cell-based | SK-MEL-5 qPCR for TRPM-1 | 0.00006-35 | 35 |
| 651772 | Cell-based | SK-MEL-5 qPCR for CDK2 | 0.00006-35 | 35 |
| 651771 | Cell-based | SK-MEL-5 qPCR for DCT | 0.00006-35 | 35 |
| 651795 | Cell-based | SK-MEL-5 qPCR for MLANA | 0.00006-35 | 35 |
| 651920 | Cell-based | Primary melanocyte cell viability | 0.00006-35 | 33 |
NA= not applicable
Appendix B. Detailed Assay Protocols
SK-MEL-5 TRPM-1 Luciferase Reporter Assay (2084-01)
SK-MEL-5/TRPM1Luc Culture Medium
- DMEM (High Glucose, HEPES, Phenol Red), Invitrogen Catalog No. 12430-047
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
- Hygromycin (250 ug/mL), Invitrogen 10687-010
SK-MEL-5/TRPM1Luc Plating Medium: DMEM (High Glucose, no Phenol Red), Invitrogen Catalog No. 31053-036, Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03, Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
Steady Glo Promega Catalog No. E2550
Parthenolide Enzo Catalog No. BML-T113-0250
SK-MEL-5/TRPM1Luc cells were maintained in DMEM (10%FBS, 1% Pen-Strep-Glutamine, 250 ug/mL Hygromycin). Cells were fluid changed every 3 days and/or split upon reaching 100% confluency. For the primary HTS, cells were thawed at 4 million cells per Falcon T175 flask. After 3 days, the cells were fluid changed. After 3 more days, the cells were passed to a Corning triple flask (10-15 million cells) and plated after 3 days in the triple flask.
Day 1
- Plate TRPM-1 luc/SKMEL5 cells at 2000 per well in 30 L media (phenol red free DMEM/10% Fetal Bovine Serum/Penicillin/Streptomycin/L-Glutamine)
- Use Corning white 384-well, square, opaque-bottomed plates (Corning Catalog No. 8867BC)
Day 2
- 3.
Pin 100 nL compound/DMSO solution (Cybi Well) into assay plates. (For HTS, required sentinel pinning with the positive control, parthenolide (6 mM))
- 4.
Incubate 24 hours at 37°C in Liconic incubator.
Day 3
- 5.
Add 20 μL 100% Promega SteadyGlo per well with Thermo Combi fluid transfer apparatus.
- 6.
Shake 15 seconds on “big bear” plate shaker and incubate at room temperature for 5 minutes.
- 7.
Read on the Perkin-Elmer EnVision plate reader with ultra-sensitive luminescence (US LUM) settings for 0.5 sec per well
SK-MEL-5 Cytotoxicity Assay (2084-02)
SK-MEL-5 Cells ATCC Catalog No. HTB-70, lot 58483232, passage 28
SK-MEL-5 Culture Medium
- DMEM (High Glucose, HEPES, Phenol Red), Invitrogen Catalog No. 12430-047
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
SK-MEL-5 Plating Medium
- DMEM (High Glucose, no Phenol Red), Invitrogen Catalog No. 31053-036
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
SK-MEL-5 cells were maintained in DMEM (10%FBS, 1% Pen-Strep-Glutamine). Cells were fluid changed every 3 days and/or split upon reaching 100% confluency. For secondary assays, cells were thawed at 4 million cells per Falcon T175 flask. After 3 days, the cells were fluid changed, after 3 more days cells were passed to a Corning Triple flask (10-15 million cells) and plated after 3 days in the triple flask.
Day 1
- Plate SK-MEL-5 cells at 3,000 per well in 30 μL media (phenol red free DMEM/10% Fetal Bovine Serum/Penicillin/Streptomycin/L-Glutamine)
- Use Corning white 384-well, square, opaque-bottomed plates (Corning Catalog No. 8867BC)
Day 2
- 3.
Pin 100 nL compound/DMSO solution (Cybi Well) into assay plates. (For HTS, required sentinel pinning with the positive control, parthenolide (6 mM))
- 4.
Incubate 24 hours at 37°C in Liconic incubator.
Day 3
- 5.
Add 20 μL 100% Cell Titer GLO per well with Thermo Combi fluid transfer apparatus.
- 6.
Shake 15 seconds on “big bear” plate shaker and incubate at room temperature for 5 minutes.
- 7.
Read on the Perkin-Elmer EnVision plate reader with luminescence (LUM) settings for 0.1 sec per well.
A-375 Cytotoxicity Assay (2084-03)
A-375 Cells ATCC Catalog No. CRL-1619, Lot No. 58463364, passage 166
A-375 Culture Medium
- DMEM (High Glucose, HEPES, Phenol Red), Invitrogen Catalog No. 12430-047
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
A-375 Plating Medium
- DMEM (High Glucose, no Phenol Red), Invitrogen Catalog No. 31053-036
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
A-375 cells were maintained in DMEM (10%FBS, 1% Pen-Strep-Glutamine). Cells were fluid changed every 3 days and/or split upon reaching 90% confluency. For secondary assays, cells were thawed at 2 million cells per Falcon T175 flask. After 3 days, cells were passed to a Corning Triple flask (6-8 million cells) and plated after 3 days in the triple flask.
Day 1
- Plate A-375 cells at 3,000 per well in 30 μL media (phenol red free DMEM/10% Fetal Bovine Serum/Penicillin/Streptomycin/L-Glutamine)
- Use Corning white 384-well, square, opaque-bottomed plates (Corning Catalog No. 8867BC)
Day 2
- 3.
Pin 100 nL compound/DMSO solution (Cybi Well) into assay plates. (For HTS, required sentinel pinning with the positive control, parthenolide (6 mM))
- 4.
Incubate 24 hours at 37°C in Liconic incubator.
Day 3
- 5.
Add 20 μL 100% Promega Cell Titer GLO per well with Thermo Combi fluid transfer apparatus.
- 6.
Shake 15 seconds on “big bear” plate shaker and incubate at room temperature for 5 minutes.
- 7.
Read on the Perkin-Elmer EnVision plate reader with luminescence (LUM) settings for 0.1 sec per well.
MALME-3M Cytotoxicity Assay (2084-04)
MALME-3M Cells ATCC Catalog No. HTB-64, Lot No. 58483222, passage 26
MALME-3M Culture Medium
- IMDM (High Glucose, Phenol Red), ATCC Catalog No. 30-2005
- Fetal Bovine Serum (20%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
MALME-3M Plating Medium
- IMDM (no Phenol Red), Gibco Catalog No. 21056-02
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
MALME-3M cells were maintained in IMDM (20%FBS, 1% Pen-Strep-Glutamine). Cells were fluid changed every 3 days and/or split upon reaching 100% confluency. For secondary assays, cells were thawed at 6 million cells per Falcon T175 flask. After 3 days, cells were fluid changed, after 3 more days cells were passed to a Corning Triple flask (15-18 million cells) and plated after 3 days in the triple flask.
Day 1
- Plate MALME-3M cells at 3,000 per well in 30 μL media (phenol red free IMDM/10% Fetal Bovine Serum/Penicillin/Streptomycin/L-Glutamine)
- Use Corning white 384-well, square, opaque-bottomed plates (Corning Catalog No. 8867BC)
Day 2
- 3.
Pin 100 nL compound/DMSO solution (Cybi Well) into assay plates, including the positive control.
- 4.
Incubate 24 hours at 37°C in Liconic incubator.
Day 3
- 5.
Add 20 μL 100% Promega Cell Titer GLO per well with Thermo Combi fluid transfer apparatus.
- 6.
Shake 15 seconds on “big bear” plate shaker and incubate at room temperature for 5 minutes.
- 7.
Read on the Perkin-Elmer EnVision plate reader with luminescence (LUM) settings for 0.1 sec per well.
Cell proliferation assay with primary human melanocytes (2084-06)
Primary human neonatal melanocytes were isolated from discarded foreskins by gentle dispase treatment and grown in TIVA media (Ham's F10 media supplemented with 7% FBS, penicillin/streptomycin/glutamine, 0.1mM IBMX, 50ng/mL TPA, 1μM Na3VO4 and 1μM dbcAMP). Cells were passaged using Accutase (Sigma Catalog #A6964-100ML) for gentle treatment and generation of a single cell suspension.
Day 1
- Plate primary melanocytes at 3,000 per well in 30 μL media (TIVA media)
- Use Corning white 384-well, square, opaque-bottomed plates (Corning Catalog No. 8867BC)
Day 2
- 3.
Pin 100 nL compound/DMSO solution (Cybi Well) into assay plates. (pinning with the positive control, parthenolide (18 μM final concentration))
- 4.
Incubate 24 hours at 37°C in Liconic incubator.
Day 3
- 5.
Add 20 μL 100% Promega Cell Titer GLO per well with Thermo Combi fluid transfer apparatus.
- 6.
Shake 15 seconds on “big bear” plate shaker and incubate at room temperature for 5 minutes.
- 7.
Read on the Perkin-Elmer EnVision plate reader with luminescence (LUM) settings for 0.1 sec per well.
qPCR Assay for Target Gene Expression (MITF: 2084-05, TRPM1: 2084-09, CDK2: 2084-11, DCT: 2084-12, MLANA: 2084-13)
SK-MEL-5 Cells ATCC Catalog No. HTB-70, lot 58483232, passage 28
SK-MEL-5 Culture/Plating Medium
- DMEM (High Glucose, HEPES, Phenol Red), Invitrogen Catalog No. 12430-047
- Fetal Bovine Serum (10%), Thermo-Hyclone Catalog No. SH30071.03
- Pen-Strep-Glutamine (1%), Invitrogen Catalog No. 10378-016
Parthenolide Enzo Catalog No. BML-T113-0250
Cells to CT Bulk Lysis Solution Ambion Catalog No. 4391851C
Cells to CT Bulk RT Reagents Ambion Catalog No. 4391852C
Light Cycler 480 Probes Master Roche Catalog No. 4887301001
Human GAPD (GAPDH) Endogenous Control VIC/MGB probe/primer limited Applied Biosystems Catalog No. 4326317E
Target Gene FAM probe/primer sets
- Human MITF FAM probe/primer Applied Biosystems Catalog No. 4331182 Hs01117294_m1
- Human TRPM1 probe/primer Applied Biosystems Catalog No. 4331182 Hs00170127_m1
- Human CDK2 probe/primer Applied Biosytems Catalog No. 4331182 Hs01548894_m1
- Human DCT probe/primer Applied Biosystems Catalog No. 4331182 Hs01098278_m1
- Human MLANA probe/primer Applied Biosystems Catalog No. 4331182 Hs00194133_m1
SK-MEL-5 cells were maintained in DMEM (10%FBS, 1% Pen-Strep-Glutamine). Cells were fluid changed every 3 days and/or split upon reaching 100% confluency. For secondary assays, cells were thawed at 4 million cells per Falcon T175 flask. After 3 days, the cells were fluid changed, after 3 more days cells were passed to a Corning triple flask (10-15 million cells) and plated after 3 days in the triple flask.
Day 1
- Plate SK-MEL-5 cells at 4,000 per well in 30 μL media (DMEM/10% Fetal Bovine Serum/Penicillin/Streptomycin/L-Glutamine)
- Use Corning white 384-well, square, opaque-bottomed plates (Corning Catalog No. 8867BC)
Day 2
- 3.
Pin 100 nL compound/DMSO solution (Cybi Well) into assay plates. (in plate positive control, parthenolide (6 mM))
- 4.
Incubate 24 hours at 37°C in Liconic incubator
Day 3
Cell Lysis
- 5.
The medium is aspirated from assay plates and the cells are washed twice (100 μL PBS) using the ELX405 Plate Washer (Biotek).
- 6.
The assay plates are flipped upside down and centrifuged at 1000 rpm for 2 minutes to remove the excess liquid.
- 7.
10 μL of Lysis solution with DNase I (Ambion, from Cell to CT Lysis Mix) is added to each well using the MultiDrop Combi/Standard tube dispensing cassette (Thermo Scientific).
- 8.
Each assay plate is then shaken for 2 minutes and incubated for an additional 8 minutes at room temperature.
- 9.
1 μL of stop solution (Ambion, from Cell to CT Lysis Mix) is added with the Multidrop Combi-nL (Thermo Scientific) and the assay plate is centrifuged at 1000 rpm for 2 minutes.
Reverse Transcription (RT) Mix
Table 1
Component Amount per reaction 2X RT Buffer 5 μL 20X RT Enzyme Mix 0.5 μL Nuclease-Free Water 2.5 μL - 10.
8 μL of RT mix is dispensed into each well of a RT assay plate (Axygen, PCR-384 RGD C).
- 11.
2 μL of the lysed cells are transferred into RT assay plate using Vario transfer unit (Cybi Well).
- 12.
The RT assay plates are incubated at 37°C for 1 hour and the reverse transcriptase is inactivated by incubating the plates for 1 minute at 95°C.
- 13.
cDNA is stored at -80°C until ready for qPCR analysis
Day 4
qPCR Master Mix
Table 2
Component Amount per reaction 2X Roche Master Mix 2.5 μL 20X FAM Target Gene Taqman probe/primer 0.125 μL 20X VIC GAPDH Taqman probe/primer 0.125 μL PCR water 1.25 μL - 14.
4 μL/well of qPCR master mix is dispensed in PCR plate (Roche Light Cycler 480 Multiwell Plate 384, Catalog No. 04 729 749 001) using the Multidrop Combi-nL (Thermo Scientific).
- 15.
1 μL/well of RT DNA is transferred in the 4 μL/well PCR plate.
- 16.
The PCR plates are centrifuged for 2 minutes at 1000 rpm.
- 17.
PCR is performed using Thermo Cycler (Roche Light Cycler 480 II) with Macro Protocol:
Step Temperature Time 1. 95°C 10 minutes 2. 95°C 10 seconds 3. 60°C 30 seconds Step 2 and 3 (55 cycles) 4. 40°C 30 seconds
Data Analysis
For the primary screen and other assays, negative-control (NC) wells and positive-control (PC) wells were included on every plate. The raw signals of the plate wells were normalized using the ‘Stimulators Minus Neutral Controls’ or the ‘Neutral Controls’ method (when no positive control was available) in GeneData Screener Assay Analyzer (v7.0.3 & v10.0.2). The median raw signal of the intra-plate NC wells was set to a normalized activity value of 0, while the median raw signal of the intra-plate PC wells was set to a normalized activity value of 100. Experimental wells were scaled to this range, resulting in an activity score representing the percent change in signal relative to the intra-plate controls. The mean of the replicate percent activities were presented as the final ‘PubChem Activity Score’. The ‘PubChem Activity Outcome’ class was assigned as described below, based on an activity threshold of 70%:
- Activity_Outcome = 1 (inactive), less than half of the replicates fell outside the threshold.
- Activity_Outcome = 2 (active), all of the replicates fell outside the threshold, OR at least half of the replicates fell outside the threshold AND the ‘PubChem Activity Score’ fell outside the threshold.
- Activity_Outcome = 3 (inconclusive), at least half of the replicates fell outside the threshold AND the ‘PubChem Activity Score did not fall outside the threshold.
Appendix C. Experimental Procedures for the Synthesis of the Probe
General synthesis and analysis experimental details: All reagents were used as received from commercial suppliers. The 1H NMR spectra were recorded on a 400 MHz Bruker Avance spectrometer equipped with a broadband observe probe or a 500 MHz Bruker AVIII spectrometer equipped with a dual cryoprobe. The 13C NMR spectra were recorded on a 500 MHz Bruker AVIII spectrometer equipped with a dual cryoprobe (at 125 MHz). Column chromatography separations were performed using the Teledyne Isco CombiFlash Rf using RediSep Rf silica gel or RediSep Rf C18 High Performance Gold columns. The analytical RPLC method used an Agilent 1200 RRLC system with UV detection (Agilent 1200 DAD SL) and mass detection (Agilent 6224 TOF). The analytical method conditions included a Waters Aquity BEH C18 column (2.1 × 50 mm, 1.7 μm) and elution with a linear gradient of 5% acetonitrile in pH 9.8 buffered aqueous ammonium formate to 100% acetonitrile at 0.4 mL/min flow rate. Compound purity was measured on the basis of peak integration (area under the curve) from UV-vis absorbance at 214 nm, and compound identity was determined on the basis of mass spectral and NMR analyses. All compounds used for biological studies have purity of ≥92%.

4-((1,4-Dioxo-1,4-dihydronaphthalen-2-yl)amino)benzenesulfonamide (ML329): Cerium chloride heptahydrate (36 mg; 97 μmol), followed by 4-aminobenzenesulfonamide (689 mg; 4.00 mmol), was added to a suspension of naphthalene-1,4-dione (316 mg; 2.00 mmol) in 95% ethanol (8.0 mL). The reaction vial was capped then put into a 75 °C block. After three days the reaction was cooled to room temperature, then 80 mL of 1.0 M citric acid was added with vigorous stirring. The insoluble material was collected by filtration, washed with water, and dried in vacuo at 45 °C affording crude product as a rust colored solid (370 mg). A portion of this (225 mg) was purified by pRPLC yielding 4-((1,4-dioxo-1,4-dihydronaphthalen-2-yl)amino)benzenesulfonamide as a rust-colored solid (101 mg).
1H NMR (400 MHz, DMSO-d6) δ 9.45 (s, 1H), 8.09 (dd, J = 1.0, 7.6 Hz, 1H), 7.98 (dd, J = 1.1, 7.6 Hz, 1H), 7.92 – 7.79 (m, 4H), 7.60 (m, 2H), 7.37 (s, 2H), 6.33 (s, 1H).
13C NMR (125 MHz, DMSO-d6) δ 183.0, 181.3, 145.2, 141.5, 139.6, 134.9, 132.9, 132.3, 130.4, 127.0, 126.2, 125.3, 122.7, 103.8.
LC-MS (ESI+): Purity at 214 nm is 100%. HRMS: 329.0591 (calcd for C16H13N2O4S = [M+H]+); 329.0594 (found / [M+H]+).
Appendix D. Experimental Procedure for Analytical Assays
Solubility. Solubility was determined in phosphate buffered saline (PBS) pH 7.4 with 1% DMSO. Each compound was prepared in duplicate at 100 μM in both 100% DMSO and PBS with 1% DMSO. Compounds were allowed to equilibrate at room temperature with a 250 rpm orbital shake for 24 hours. After equilibration, samples were analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. The DMSO samples were used to create a two-point calibration curve to which the response in PBS was fit.
PBS Stability. Stability was determined in the presence of PBS pH 7.4 with 0.1% DMSO. Each compound was prepared in duplicate on six separate plates and allowed to equilibrate at room temperature with a 250-rpm orbital shake for 48 hours. One plate was removed at each time point (0, 2, 4, 8, 24, and 48 hours). An aliquot was removed from each well and analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. Additionally, to the remaining material at each time point, acetonitrile was added to force dissolution of compound (to test for recovery of compound). An aliquot of this was also analyzed by UPLC-MS.
GSH Stability. Stability was determined in the presence of PBS pH 7.4, 10 μM compound and 50 μM glutathione with 0.1% DMSO. Each compound was prepared in duplicate on six separate plates and allowed to equilibrate at room temperature with a 250-rpm orbital shake for 48 hours. One plate was removed at each time point (0, 2, 4, 8, 24, and 48 hours). An aliquot was removed from each well and analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. Additionally, to the remaining material at each time point, acetonitrile was added to force dissolution of compound (to test for recovery of compound). An aliquot of this was also analyzed by UPLC-MS.
DTT Stability. Compound was dissolved at 10 μM in PBS/acetonitrile (1/1) at pH 7.4 (1% DMSO) and incubated at room temperature with either no thiol source as a negative control or 50 μM dithiothreitol (DTT). The mixtures were sampled every hour for eight hours or every 8 hours for 48 hours and analyzed by RP HPLC/UV/HRMS. The analytical RP HPLCUV/HRMS system utilized for the analysis was a Waters Acquity system with UV-detection and mass-detection (Waters LCT Premier). The analytical method conditions included a Waters Acquity HSS T3 C18 column (2.1 × 50mm, 1.8um) and elution with a linear gradient of 99% water to 100% CH3CN at 0.6 mL/min flow rate. Peaks on chromatograms were integrated using the Waters OpenLynx software. Absolute areas under the curve (214 nm) were compared at each time point to determine relative percent compound remaining in supernatant. The masses of potential adducts were searched for in the samples to determine if any detectable adduct formed. All samples were prepared in duplicate. Ethacrynic acid, a known Michael acceptor, was used as a positive control and was tested in PBS/acetonitrile (1/1).
Plasma Protein Binding. Plasma protein binding was determined by equilibrium dialysis using the Rapid Equilibrium Dialysis (RED) device (Pierce Biotechnology, Rockford, IL) for both human and mouse plasma. Each compound was prepared in duplicate at 5 μM in plasma (0.95% acetonitrile, 0.05% DMSO) and added to one side of the membrane (200 μL) with PBS pH 7.4 added to the other side (350 μL). Compounds were incubated at 37 °C for 5 hours with a 250-rpm orbital shake. After incubation, samples were analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer.
Plasma Stability. Plasma stability was determined at 37°C at 5 hours in both human and mouse plasma. Each compound was prepared in duplicate at 5 μM in plasma diluted 50/50 (v/v) with PBS pH 7.4 (0.95% acetonitrile, 0.05% DMSO). Compounds were incubated at 37°C for 5 hours with a 250-rpm orbital shake with time points taken at 0 hours and 5 hours. Samples were analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer.
Appendix E. Chemical Characterization Data for Probe
Appendix F. Chemical Characterization Data for All Analogs
Appendix G. Prior Art Search
Appendix H. Eurofins Panlabs LeadProfiling Screen Report for ML329
The following text was provided along with the study results in the report for the LeadProfilingScreen for ML329.
Study Objective
To evaluate, in radioligand binding assays, the activity of probe compound ML329 (KUC111774N-02) across a panel of 67 receptors.
Methods
Methods employed in this study have been adapted from the scientific literature to maximize reliability and reproducibility. Reference standards were run as an integral part of each assay to ensure the validity of the results obtained.
Where presented, IC50 values were determined by a non-linear, least squares regression analysis using MathIQ™ (ID Business Solutions Ltd., UK). Where inhibition constant (Ki) are presented, the Ki values were calculated using the equation of Cheng and Prusoff (Cheng. Y., Prusoff, W.H., Biochem. Pharmacol. 22:3099-3108, 1973) using the observed IC50 of the tested compound, the concentration of radioligand employed in the assay, and the historical values of the KD of the ligand (obtained experimentally at Eurofins Panlabs, Inc.). Where presented, the Hill coefficient (nH), defining the slope of the competitive binding curve, was calculated using MathIQ™. Hill coefficients significantly different than 1.0, may suggest that the binding displacement does not follow the laws of mass action with a single binding site. Where IC50, Ki, and/or nH data are presented without Standard Error of the Mean (SEM), data are insufficient to be quantitative, and the values presented (Ki, IC50, nH) should be interpreted with caution.
| Cat # | Assay Name | Batch* | Spec. | Rep. | Conc. | % Inh. |
|---|---|---|---|---|---|---|
| 200510 | Adenosine A1 | 324045 | human | 2 | 10 μM | 46 |
| 200610 | Adenosine A2A | 324044 | human | 2 | 10 μM | 50 |
| 200720 | Adenosine A3 | 324012 | human | 2 | 10 μM | 15 |
| 203100 | Adrenergic α1A | 324046 | rat | 2 | 10 μM | 28 |
| 203200 | Adrenergic α1B | 324024 | rat | 2 | 10 μM | 16 |
| 203400 | Adrenergic α1D | 324047 | human | 2 | 10 μM | 21 |
| 203620 | Adrenergic α2A | 324048 | human | 2 | 10 μM | 33 |
| 204010 | Adrenergic β1 | 324027 | human | 2 | 10 μM | 20 |
| 204110 | Adrenergic β2 | 324042 | human | 2 | 10 μM | 10 |
| 285010 | Androgen (Testosterone) AR | 324064 | rat | 2 | 10 μM | 14 |
| 212510 | Bradykinin B1 | 324141 | human | 2 | 10 μM | 29 |
| 212620 | Bradykinin B2 | 324310 | human | 2 | 10 μM | 11 |
| 214510 | Calcium Channel L-Type, Benzothiazepine | 324113 | rat | 2 | 10 μM | 20 |
| 214600 | Calcium Channel L-Type, Dihydropyridine | 324317 | rat | 2 | 10 μM | 30 |
| 216000 | Calcium Channel N-Type | 323997 | rat | 2 | 10 μM | -3 |
| 217030 | Cannabinoid CB1 | 324041 | human | 2 | 10 μM | 31 |
| 219500 | Dopamine D1 | 324037 | human | 2 | 10 μM | 21 |
| 219700 | Dopamine D2S | 324038 | human | 2 | 10 μM | 0 |
| 219800 | Dopamine D3 | 324039 | human | 2 | 10 μM | 9 |
| 219900 | Dopamine D4.2 | 324230 | human | 2 | 10 μM | 4 |
| 224010 | Endothelin ETA | 324347 | human | 2 | 10 μM | 17 |
| 224110 | Endothelin ETB | 324349 | human | 2 | 10 μM | 17 |
| 225510 | Epidermal Growth Factor (EGF) | 324350 | human | 2 | 10 μM | 11 |
| 226010 | Estrogen ERα | 324351 | human | 2 | 10 μM | 5 |
| 226600 | GABAA, Flunitrazepam, Central | 324035 | rat | 2 | 10 μM | 20 |
| 226500 | GABAA, Muscimol, Central | 324034 | rat | 2 | 10 μM | 4 |
| 228610 | GABAB1A | 324354 | human | 2 | 10 μM | 11 |
| 232030 | Glucocorticoid | 324040 | human | 2 | 10 μM | 5 |
| 232700 | Glutamate, Kainate | 324324 | rat | 2 | 10 μM | 43 |
| 232810 | Glutamate, NMDA, Agonism | 324326 | rat | 2 | 10 μM | 29 |
| 232910 | Glutamate, NMDA, Glycine | 324329 | rat | 2 | 10 μM | 9 |
| 233000 | Glutamate, NMDA, Phencyclidine | 324036 | rat | 2 | 10 μM | 0 |
| 239610 | Histamine H1 | 324052 | human | 2 | 10 μM | -5 |
| 239710 | Histamine H2 | 324083 | human | 2 | 10 μM | -5 |
| 239820 | Histamine H3 | 324131 | human | 2 | 10 μM | -5 |
| 241000 | Imidazoline I2, Central | 324053 | rat | 2 | 10 μM | -5 |
| 243520 | Interleukin IL-1 | 324013 | mouse | 2 | 10 μM | 6 |
| 250460 | Leukotriene, Cysteinyl CysLT1 | 324130 | human | 2 | 10 μM | 5 |
| 251600 | Melatonin MT1 | 324356 | human | 2 | 10 μM | 9 |
| 252610 | Muscarinic M1 | 324054 | human | 2 | 10 μM | 7 |
| 252710 | Muscarinic M2 | 324055 | human | 2 | 10 μM | 15 |
| 252810 | Muscarinic M3 | 324056 | human | 2 | 10 μM | 22 |
| 257010 | Neuropeptide Y Y1 | 324363 | human | 2 | 10 μM | -6 |
| 257110 | Neuropeptide Y Y2 | 324365 | human | 2 | 10 μM | 5 |
| 258590 | Nicotinic Acetylcholine | 324018 | human | 2 | 10 μM | 9 |
| 258700 | Nicotinic Acetylcholine α1, Bungarotoxin | 324019 | human | 2 | 10 μM | -2 |
| 260130 | Opiate δ1 (OP1, DOP) | 324366 | human | 2 | 10 μM | -6 |
| 260210 | Opiate κ(OP2, KOP) | 324124 | human | 2 | 10 μM | 19 |
| 260410 | Opiate μ(OP3, MOP) | 324058 | human | 2 | 10 μM | 1 |
| 264500 | Phorbol Ester | 324067 | mouse | 2 | 10 μM | 18 |
| 265010 | Platelet Activating Factor (PAF) | 324114 | human | 2 | 10 μM | 3 |
| 265600 | Potassium Channel [KATP] | 324068 | hamster | 2 | 10 μM | 10 |
| 265900 | Potassium Channel hERG | 324069 | human | 2 | 10 μM | -4 |
| 268420 | Prostanoid EP4 | 324070 | human | 2 | 10 μM | 11 |
| 268700 | Purinergic P2X | 324103 | rabbit | 2 | 10 μM | -2 |
| 268810 | Purinergic P2Y | 324128 | rat | 2 | 10 μM | -7 |
| 270000 | Rolipram | 324059 | rat | 2 | 10 μM | 15 |
| 271110 | Serotonin (5-Hydroxytryptamine) 5-HT1A | 324121 | human | 2 | 10 μM | 21 |
| 271700 | Serotonin (5-Hydroxytryptamine) 5-HT2B | 324061 | human | 2 | 10 μM | 37 |
| 271910 | Serotonin (5-Hydroxytryptamine) 5-HT3 | 324005 | human | 2 | 10 μM | 5 |
| 278110 | Sigma σ1 | 324062 | human | 2 | 10 μM | 15 |
| 279510 | Sodium Channel, Site 2 | 324063 | rat | 2 | 10 μM | 6 |
| 255520 | Tachykinin NK1 | 324358 | human | 2 | 10 μM | 37 |
| 285900 | Thyroid Hormone | 324137 | rat | 2 | 10 μM | -4 |
| 220320 | Transporter, Dopamine (DAT) | 324015 | human | 2 | 10 μM | 2 |
| 226400 | Transporter, GABA | 324353 | rat | 2 | 10 μM | 9 |
| 204410 | Transporter, Norepinephrine (NET) | 324014 | human | 2 | 10 μM | 8 |
| 274030 | Transporter, Serotonin (5-Hydroxytryptamine) (SERT) | 324370 | human | 2 | 10 μM | -5 |
- *
Batch: Represents compounds tested concurrently in the same assay(s).
Appendix I. Primary melanocyte viability assay data
Figure A.1MLS004556029 (CID 12387471, SID 144221520) was tested across a range of concentrations up to 35 μM in a primary human melanocyte assay
Compounds were incubated for 24 hours and viability measured with CellTiter-Glo (AID 651920). Concentration response curves were generated with Genedata Screener Condeseo and show normalized percent activity for the individual doses. IC50 = 7.14 μM. ○=replicate 1, △=replicate 2, ◻=replicate 3
Appendix J. Compounds Provided to Evotec
Table A3Probe and Analog Information
| CID / SID | Broad ID / KU ID | P/A | MLSID | ML |
|---|---|---|---|---|
| 12387471 / 144221520 | BRD-K73037408-001-01-5 / KUC111774N-03 | P | MLS004556029 | ML329 |
| 81124 / 144221523 | BRD-K29842115-001-07-4 / KUC111337N-02 | A | MLS004556032 | NA |
| 72909 / 144221522 | BRD-K75502546-001-01-9 / KUC111359N-02 | A | MLS004556031 | NA |
| 279009 / 144221524 | BRD-K48101260-001-01-8 / KUC111761N-02 | A | MLS004556033 | NA |
| 56951846 / 144221521 | BRD-K93744531-001-01-2 / KUC111358N-02 | A | MLS004556030 | NA |
| 56928031 / 135611193 | BRD-K16604218-001-01-2 / KUC111109N | A | MLS004556034 | NA |
A = analog; NA= not applicable; P = probe
- Probe Structure & Characteristics
- Recommendations for scientific use of the probe
- Introduction
- Materials and Methods
- Results
- Discussion
- References
- Assay Summary Table
- Detailed Assay Protocols
- Experimental Procedures for the Synthesis of the Probe
- Experimental Procedure for Analytical Assays
- Chemical Characterization Data for Probe
- Chemical Characterization Data for All Analogs
- Prior Art Search
- Eurofins Panlabs LeadProfiling Screen Report for ML329
- Primary melanocyte viability assay data
- Compounds Provided to Evotec
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- A Small Molecule Inhibitor of the MITF Molecular Pathway - Probe Reports from th...A Small Molecule Inhibitor of the MITF Molecular Pathway - Probe Reports from the NIH Molecular Libraries Program
- Identification of Functionally Selective Small Molecule Antagonists of the Neuro...Identification of Functionally Selective Small Molecule Antagonists of the Neuropeptide-S Receptor: Naphthopyranopyrimidines - Probe Reports from the NIH Molecular Libraries Program
- Identification of small molecules that selectively inhibit streptokinase express...Identification of small molecules that selectively inhibit streptokinase expression without suppression of viability in Group A streptococci - Probe 2 - Probe Reports from the NIH Molecular Libraries Program
- Optimization and characterization of a pan protein arginine deiminase (PAD) inhi...Optimization and characterization of a pan protein arginine deiminase (PAD) inhibitor - Probe Reports from the NIH Molecular Libraries Program
- Identification of Small Molecule Antagonists of the Neuropeptide-S Receptor - Pr...Identification of Small Molecule Antagonists of the Neuropeptide-S Receptor - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...