NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Type Ia Congenital Disorders of Glycosylation (CDG-Ia) is the most common form of the Congenital Disorders of Glycosylation. This autosomal recessive disorder impairs the synthesis of N-linked oligosaccharide chains and is caused by defects in the PMM2 gene. PMM2 encodes phosphomannomutase 2, which is responsible for the conversion of mannose-6-P [Man-6-P] to Man-1-P. Currently, there is no treatment for CDG-Ia patients. The current project aimed to identify novel non-competitive inhibitors of phosphomannose isomerase, PMI. as potential therapeutic treatments for these patients. The developed probe ML096 (CID-25199533) inhibits human PMI and may inhibit other PMI orthologs due to the highly conserved nature of the enzyme. The probe is membrane permeable and, as a result can also be used to inhibit PMI in living cells. Thus, the probe may serve as a therapeutic treatment for CDG-Ia patients by blocking the catabolism of Man-6-P in cells, thereby redirecting it towards protein glycosylation using this residual PMM2 activity.
Assigned Assay Grant #: R03 MH082386-01
Screening Center Name & PI: Conrad Prebys Center for Chemical Genomics (formerly Burnham Center for Chemical Genomics) & Dr. John C. Reed
Chemistry Center Name & PI: Conrad Prebys Center for Chemical Genomics (formerly Burnham Center for Chemical Genomics) & Dr. John C. Reed
Assay Submitter & Institution: Dr. Hudson Freeze, Sanford-Burnham Medical Research Institute (formerly Burnham Institute for Medical Research)
PubChem Summary Bioassay Identifier (AID): AID-1545
Probe Structure & Characteristics

| CID/ML | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Selectivity | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID-25199533 ML096 | Phosphomannose Isomerase (PMI) | 1,070 nM IC50 SID-57309177
AID-1535 | PMM2 | >100 µM IC50 SID-57309177 AID-1655 | >93 X |
Recommendations for the scientific use of this probe
The probe can be used to inhibit human phosphomannose isomerase and due to the high homology between other eukaryotic orthologs, it will likely inhibit them as well. The probe can also be used to inhibit this enzyme in living cells since it is membrane permeable. One use of the probe would be to block catabolism of mannose-6-P in cells and direct it toward protein glycosylation. It may have therapeutic potential in phosphomannomutase 2-deficient CDG-Ia patients who are given mannose dietary supplements.
1. Scientific Rationale for Project
Congenital Disorders of Glycosylation (CDG) are autosomal recessive defects in the synthesis of N-linked oligosaccharide chains (1). CDG group I (CDG-I) defects are defined as those caused by mutations in genes encoding enzymes used for the synthesis and transfer of lipid linked oligosaccharide (LLO) to newly synthesized proteins in the lumen of the ER. The steps in this pathway and the genes encoding them are very similar from yeast to human. It requires 30–40 single gene products, each dependent on the previous step in the linear sequence to produce and transfer the LLO to protein. Therefore, mutations in any step may cause a type of CDG. There is considerable overlap in the clinical presentations between different types of CDG and a broad diversity within each type. The most common form of CDG, called Type Ia (CDG-Ia), is caused by defects in PMM2 (Man-6-P to Man-1-P), the gene that encodes phosphomannomutase. Mortality is 20% in the first 5 years, but then patients stabilize. Currently, there is no treatment for the CDG-Ia. CDG-Ib patients, who are deficient in phosphomannose isomerase (PMI) catalyzing conversion of Man-6-P to Fru-6-P, are successfully treated with free mannose (2 – 6). Unfortunately, mannose therapy is not effective for CDG-Ia patients, most likely due to efficient Man-6-P consumption in the PMI reaction (7,8). It is believed that patients with Congenital Disorder of Glycosylation Type Ia (CDG-Ia) will benefit from dietary mannose if there is a simultaneous reduction of phosphomannose isomerase (PMI) activity. This would allow a modest intracellular accumulation of Man-6-P and drive metabolic flux into the glycosylation pathway using the residual PMM2 activity (9). It is assumed that a non-competitive inhibitor would work best in this setting; however, identification of chemical probes with diverse modes of action (MOA) would be advantageous for further characterization of PMI and PMM variants.
We proposed that CDG-Ia patients would benefit from dietary mannose if we simultaneously reduce PMI activity with a non-competitive or un-competitive inhibitor. This would allow a modest intracellular accumulation of Man-6-P and drive metabolic flux into the glycosylation pathway using the residual PMM2 activity.
Novel chemical probes elucidated in this way are invaluable tools to help explain the role of PMI in various biochemical pathways, and will ultimately lead to the first therapy for the growing number of CDG-Ia patients.
2. Project Description
a. The original goal for probe characteristics
The original overall goal for this project was to identify novel non-competitive inhibitors of phosphomannose isomerase (PMI) that can be used as a therapeutic for treating patients with Congenital Disorder of Glycosylation Type Ia (CDG-Ia). A non-competitive inhibitor is preferred (yet not required) to avoid competition by the increased Man-6-P level. The main aims undertaken were: 1) Identify small molecule compounds in the MLSMR collection screening 2) Validated the specificity, activity, stability and kinetic behavior of the selected compound(s) 3) Test selected inhibitors of PMI for in cell based assay to determine their effects on glycosylation in intact cells.
b. Assay implementation and screening
i. PubChem Bioassay Name(s), AID(s), Assay-Type (Primary, DR, Counterscreen, Secondary)
| PubChemBioAssay Name | AIDs | Probe Type | Assay Type | Assay Format | Assay Detection & well format |
|---|---|---|---|---|---|
| HTS identification of compounds inhibiting phosphomannose isomerase (PMI) via a fluorescence intensity assay | 1209 | Inhibitor | Primary/Confirmatory | Biochemical | Fluorescence 1536 well |
| HTS identification of compounds inhibiting phosphomannose isomerase (PMI) via a fluorescence intensity assay using a high concentration of mannose 6-phosphate [Confirmatory] | 1220 | Inhibitor | Primary/Confirmatory/Mechanistic | Biochemical | Fluorescence 1536 well |
| Confirmation of compounds inhibiting phosphomannose isomerase (PMI) via a fluorescence intensity assay. [Confirmatory] | 1535 | Inhibitor | SAR | Biochemical | Fluorescence 384 well |
| Confirmation of compounds inhibiting phosphomannose isomerase (PMI) via a fluorescence intensity assay using a high concentration of mannose 6-phosphate. [Confirmatory] | 1536 | Inhibitor | SAR/mechanistic | Biochemical | Fluorescence 384 well |
| uHTS Identification of Diaphorase Inhibitors and Chemical Oxidizers: Counter Screen for Diaphorase-based Primary Assays [Primary Screening] | 1217 | Inhibitor | Primary/Counter screen | Biochemical | Fluorescence 1536 well |
| Counter Screen for Glucose-6-Phosphate Dehydrogenase-based Primary Assay | 1020 | Inhibitor | Primary/Counter Screen | Biochemical | Fluorescence 1536 well |
| Phosophomannose Mutase 2 (PMM2) | 1655 | Inhibitor | Confirmatory/Counter screen | Biochemical | Fluorescence 1536 well |
ii. Assay Rationale & Description
The purpose of this assay was to identify inhibitors of human PMI. This was accomplished by using a G6PD-NADPH-coupled assay. In the assay PMI activity was detected through conversion of its product, fructose-6-phosphate, to glucose-6-phosphate catalyzed by phosphoglucose isomerase (PGI) and subsequent oxidation of glucose-6-phosphate to 6-phosphogluconolactone concomitant with NADP-to-NADPH conversion catalyzed by glucose-6-phosphate dehydrogenase (G6PDH). The NADPH was then detected via a resazurin-diaphorase fluorogenic reaction.
This assay was performed in the presence of near-Km concentrations of the PMI substrate, mannose-6-phosphate, to ensure the identification of inhibitors with diverse Mechanism of Actions (MOAs). Keeping in mind that non-or un-competitive inhibitors might provide a better probe, we also developed and utilized for the full-deck screening the assay performed in the presence of 10xKm of the PMI substrate be better for to help with prioritization of hits for follow-up work. The idea being, that the competitive inhibitors would show much less inhibition in these conditions comparing to the normal screening. Thus, we selected scaffolds that inhibit in both assays with similar potency for further work; the described herein probe originated from this type of scaffold. In addition, we also developed and implemented the following assays for counter screening: G6PDH (primary HTS), diaphorase (primary HTS). Compounds inhibitory in any of these counter-screen assays were excluded from further consideration.
Protocol
PMI assay materials
- Human PMI protein was provided by Dr. Hudson Freeze (Burnham Institute for Medical Research, San Diego, CA). The target protein, recombinant Phosphomannose Isomerase (PMI), was made in 1 L batches from high expressing baculovirus systems in SF9 insect cells in Dr Hudson Freeze laboratory.
- Substrate working solution: 50 mM HEPES, pH 7.4, 0.4 mM Mannose-6-phosphate, 1.6 U/ml Diaphorase, 0.2 mM Resazurin.
- Enzyme working solution: 50 mM HEPES, pH 7.4, 0.44 mM NADP+, 9.048 mM MgCl2, 0.01% Tween 20, 4.6 ug/ml phosphoglucose isomerase, 30 ng/ml PMI, 1.8 ug/ml G6PDH.
Table of Reagents and source used in experiments
| Reagents name | Vendors name |
|---|---|
| PhosphoGlucose Isomerase (PGI) | Roche Diagnostics |
| Glucose-6 phosphate Dehydrogenase (G6PD) | USB |
| Mannose-6 Phosphate | Sigma-Aldrich |
| NADP | ISC/Bioexpress |
| Hepes Buffer | Sigma-Aldrich |
| Magnesium Chloride | Sigma-Aldrich |
PMI HTS protocol
- 2 uL of Substrate working solution was added to columns 3–48 of a Costar 1536- well black plate (cat #3724) using a Thermo Multidrop Combi dispenser
- 2 ul of Substrate working solution without mannose-6-p was added to columns 1 and 2 (positive control) of a Costar 1536-well black plate (cat #3724) using a Thermo Multidrop Combi dispenser
- 40 nL of 100% DMSO was added to columns 1–4 using a HighRes biosolutions pintool and V&P Scientific pins
- 40 nL of 2 mM compounds in 100% DMSO were dispensed in columns 5–48 using a HighRes biosolutions pintool and V&P Scientific pins
- 2 uL of Enzyme working solution was added to the whole plate using a Thermo Multidrop Combi dispenser.
- Plates were incubated at room temperature for 20 min.
- After 20 minutes the plates were read on a ViewLux plate reader (Perkin Elmer), Ex544, Em590.
- The screening was performed using a HighRes biosolution fully integrated HTS POD-based system
- Data analysis was performed using CBIS software (ChemInnovations, Inc). Compounds showing more than 50% inhibition were requested from MLSMR for dose-response confirmation.
PMI dose-response assay protocol
- 9 uL of Substrate working solution was added to columns 3–24 of a Greiner 384-well black plates (cat #784076) using a Thermo Multidrop Combi dispenser
- 9 ul of Substrate working solution without mannose-6-p was added to columns 1 and 2 (positive control) using a Thermo Multidrop Combi dispenser
- 2 uL of serially diluted compounds in 10% DMSO were added to columns 3–22 using Biomek FX 3) 2 uL of 10% DMSO were added to columns 1–2, 23–24
- 9 uL of Enzyme working solution was added to the whole plate using a Thermo Multidrop Combi dispenser.
- Plates were incubated at room temperature for 20 min.
- After 20 minutes the plates were read on an Analyst plate reader (Molecular Devices), Ex544, Em590.
- Data analysis was performed using CBIS software (ChemInnovations, Inc).
iii. Summary of Results
A total of 194,158 compounds were tested at 20µM and 926 had an activity ≥ 50% in the assay. The average Z’ for the screen was 0.81, signal to background was 3.9, signal to noise was 52.4 and signal window was 19.1. Analog-by-catalog (ABC) SAR studies on the dithiazolimine series are summarized in Table 1 below.
Table 1
In vitro enzyme activity & SAR for dithiazolimine PMI inhibitors.
c. Probe Optimization
i. SAR & chemistry strategy (including structure and data) that led to the probe
Development of initial SAR around the dithiazolimine series of PMI inhibitors via purchased and synthesized analogues established 3 structural features critical for the inhibitory activity of CID-25199533 (MLS-0390940) and its derivatives: (a) a dithiazole core ring, (b) a pendant aromatic or heteroaromatic ring, and (c) a chlorine in the 5 position of the dithiazole ring. These required features are exemplified in Figure 1. Systematic replacement of each of these key moieties resulted in derivates having significantly diminished inhibitory activity (examples not shown). Thus, having established these required features, further SAR optimization efforts focused on two main goals: (1) development of inhibitors with increased potency in vitro against PMI and (2) optimization of the selectivity of these potent inhibitors over PMM. The in vitro enzyme inhibition and cell-based efficacy data for selected analogues of CID-25199533 (MLS-0390940) are shown in Table 1. Compound CID-715715 (MLS-0053966) was the primary lead compound based on the initial in vitro HTS data. This compound showed acceptable PMI inhibition (IC50 = 2.91 µM) and selectivity over PMM (IC50 >100 µM), thus analogue synthesis was executed around this hit. A variety of analogues were prepared to elucidate the optimal substitutions of the aryl ring. As can be seen in Table 1, methoxy substituents as in CID-602897 (MLS-0390826) and CID-483894 (MLS-0390822) suggested that the presence of hydrogen-bonding moieties were essential for activity. A key discovery was made when ortho nitro substituents were incorporated, yielding potent analogues such as CID-3919997 (MLS-0390820), which had acceptable potency (PMI IC50 = 3.52 µM). However, this analogue showed slight inhibition of PMM (IC50 = 58.4 µM). A companion series having a heteroaromatic substituent in place of the phenyl ring was showing potent PMI inhibition and PMM selectivity, as exemplified by the thiadiazole-containing analogue CID-6114286 (MLS-0390818; PMI IC50 = 2.3 µM; PMM IC50 > 100 µM). It was envisioned that this replacement represented an opportunity to maintain PMI potency while improving selectivity over PMM. Thus, selected analogues were synthesized incorporating small heteroaryl rings at this position, yielding potent and selective derivatives such as CID-5527821 (MLS-0390816) (PMI IC50 = 4.34 µM; PMM IC50 > 100 µM) and the probe compound CID-25199533 (MLS-0390940), a potent and selective PMI inhibitor with IC50 = 1.07 µM and completely inactive at PMM when tested at concentrations up to 100 µM.
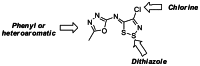
Figure 1
Summary of required features for CID-25199533 and analogues.
3. Probe
a. Chemical name
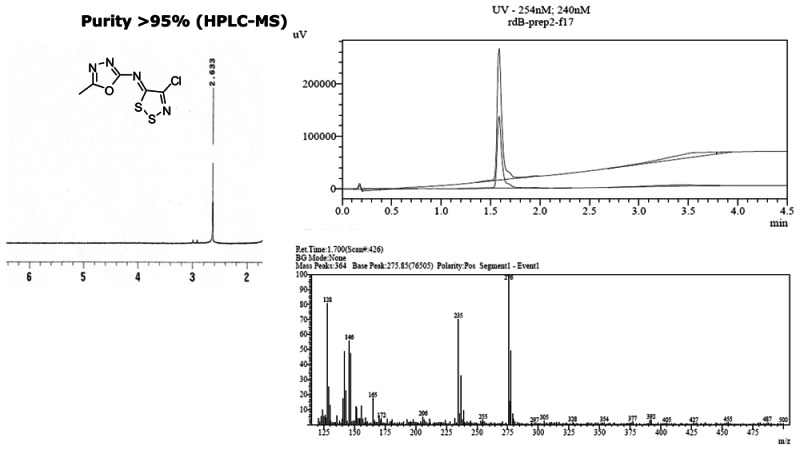
4-chloro-N-(5-methyl-1,3,4-oxadiazol-2-yl)dithiazol-5-imine [ML096]
b. Probe chemical structure
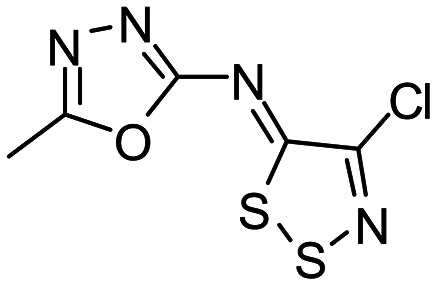
d. PubChem CID (corresponding to the SID)
CID-25199533
e. Availability from a vendor
This probe is not commercially available.
f. MLS#'s of probe molecule and five related samples that were submitted to the SMR collection
| Probe/Analog | MLS-# (BCCG#) | CID | SID | Source (vendor or BCCG syn) | Amt (mg) | Date ordered/submitted |
|---|---|---|---|---|---|---|
| Probe | MLS-0390940 | 25199533 | 57309177 | BCCG syn | 15 | 5/18/09 |
| Analog 1 | MLS-0390932 | 25181240 | 57287676 | BCCG syn | 15 | 5/18/09 |
| Analog 2 | MLS-0053966 | 715715 | 4245711 | Chemical Block | 15 | 5/18/09 |
| Analog 3 | MLS-0390826 | 602897 | 57287569 | Chemical Block | 15 | 5/18/09 |
| Analog 4 | MLS-0390818 | 6114286 | 57287561 | BCCG syn | 15 | 5/18/09 |
| Analog 5 | MLS-0390829 | 715698 | 57287572 | BCCG syn | 15 | 5/18/09 |
g. Mode of action for biological activity of probe
The mode of action is not yet fully characterized. The dithiazolimine scaffold with all its primary hits was identified as non- or un-competitive since it inhibited irrespective of substrate concentration. As alterations to the structure of the primary hits that were made to arrive at the probe are limited, the probe is very likely to have the same MOA.
h. Detailed synthetic pathway for making probe
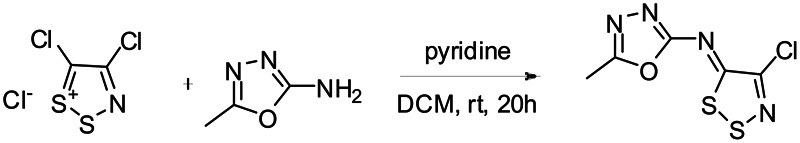
Scheme 1Synthesis of CID-25199533
i. Probe properties (solubility, absorbance/fluorescence, reactivity, toxicity, etc.)
CID-25199533 (MLS-0390940) demonstrated potent inhibition and also demonstrated selectivity within the class.
j. Properties Computed from Structure
| Molecular Weight | 234.68652 |
| Molecular Formula | C5H3ClN4OS2 |
| XLogP3-AA | 2.1 |
| H-Bond Donor | 0 |
| H-Bond Acceptor | 5 |
| Rotatable Bond Count | 1 |
| Exact Mass | 233.94368 |
| Topological Polar Surface Area | 114 |
| Heavy Atom Count | 13 |
| Formal Charge | 0 |
4. Appendices
a. Comparative data on (1) probe, (2) similar compound structures (establishing SAR) and (3) prior probes
See list of compounds ordered for SAR by catalog attached separately to end of this probe report.
b. Comparative data showing probe specificity for target
See Table 1 above for selectivity
5. Bibliography
- 1.
- Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–551. [PubMed: 16755287]
- 2.
- Alper J. Searching for medicine’s sweet spot. Science. 2001;291:2338–2343. [PubMed: 11269308]
- 3.
- Niehues R, Hasilik M, Alton G, Körner C, Schiebe-Sukumar M, Koch HG, Zimmer KP, Wu R, Harms E, Reiter K, von Figura K, Freeze HH, Harms HK, Marquardt T. Carbohydratedeficient glycoprotein syndrome type Ib. Phosphomannose isomerase deficiency and mannose therapy. J Clin Invest. 1998;101:1414–1420. [PMC free article: PMC508719] [PubMed: 9525984]
- 4.
- Babovic-Vuksanovic D, Patterson MC, Schwenk WF, O’Brien JF, Vockley J, Freeze HH, Mehta DP, Michels VV. Severe hypoglycemia as a presenting symptom of carbohydrate deficient glycoprotein syndrome. J Pediatr. 1999;135:775–781. [PubMed: 10586187]
- 5.
- de Lonlay P, Cuer M, Vuillaumier-Barrot S, Beaune G, Castelnau P, Kretz M, Durand G, Saudubray JM, Seta N. Hyperinsulinemic hypoglycemia as a presenting sign in phosphomannose isomerase deficiency: A new manifestation of carbohydrate-deficient glycoprotein syndrome treatable with mannose. J Pediatr. 1999;135:379–383. [PubMed: 10484808]
- 6.
- Westphal V, Kjaergaard S, Davis JA, Peterson SM, Skovby F, Freeze HH. Genetic and metabolic analysis of the first adult with congenital disorder of glycosylation type ib: long-term outcome and effects of mannose supplementation. Mol Genet Metab. 2001;73:77–85. [PubMed: 11350186]
- 7.
- Rush JS, Panneerselvam K, Waechter CJ, Freeze HH. Mannose supplementation corrects GDP-mannose deficiency in cultured fibroblasts from some patients with Congenital Disorders of Glycosylation (CDG). Glycobiology. 2000;10:829–835. [PubMed: 10929009]
- 8.
- Panneerselvam K, Freeze HH. Mannose corrects altered N-glycosylation in carbohydratedeficient glycoprotein syndrome fibroblasts. J Clin Invest. 1996;97:1478–1487. [PMC free article: PMC507208] [PubMed: 8617881]
- 9.
- Freeze HH. Chapter 6: Monosaccharide Metabolism. In: Varki A, Cummings R, Esko JD, Freeze HH, Hart G, Marth JD, editors. Essentials of Glycobiology. New York: Cold Spring Harbor Laboratory Press; 1999. pp. 69–84.
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Therapeutic Inhibitors of Phosphomannose Isomerase - Probe 1.[Probe Reports from the NIH Mol...]Review Therapeutic Inhibitors of Phosphomannose Isomerase - Probe 1.Hedrick M, Brown B, Rascon J, Sergienko E, Sharma V, Ng B, Ichikawa M, Freeze H, Stonich D, Su Y, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Phosphomannose isomerase inhibitors improve N-glycosylation in selected phosphomannomutase-deficient fibroblasts.[J Biol Chem. 2011]Phosphomannose isomerase inhibitors improve N-glycosylation in selected phosphomannomutase-deficient fibroblasts.Sharma V, Ichikawa M, He P, Scott DA, Bravo Y, Dahl R, Ng BG, Cosford ND, Freeze HH. J Biol Chem. 2011 Nov 11; 286(45):39431-8. Epub 2011 Sep 26.
- Potent, selective, and orally available benzoisothiazolone phosphomannose isomerase inhibitors as probes for congenital disorder of glycosylation Ia.[J Med Chem. 2011]Potent, selective, and orally available benzoisothiazolone phosphomannose isomerase inhibitors as probes for congenital disorder of glycosylation Ia.Dahl R, Bravo Y, Sharma V, Ichikawa M, Dhanya RP, Hedrick M, Brown B, Rascon J, Vicchiarelli M, Mangravita-Novo A, et al. J Med Chem. 2011 May 26; 54(10):3661-8. Epub 2011 May 3.
- Review [Congenital disorders of glycosylation].[Ann Pharm Fr. 2003]Review [Congenital disorders of glycosylation].Durand G, Dupré T, Vuillaumier-Barrot S, Seta N. Ann Pharm Fr. 2003; 61(5):330-9.
- N-glycosylation deficiency reduces ICAM-1 induction and impairs inflammatory response.[Glycobiology. 2014]N-glycosylation deficiency reduces ICAM-1 induction and impairs inflammatory response.He P, Srikrishna G, Freeze HH. Glycobiology. 2014 Apr; 24(4):392-8. Epub 2014 Jan 28.
- Therapeutic Inhibitors of Phosphomannose Isomerase - Probe 2 - Probe Reports fro...Therapeutic Inhibitors of Phosphomannose Isomerase - Probe 2 - Probe Reports from the NIH Molecular Libraries Program
- Re-evaluation of Some Organic Chemicals, Hydrazine and Hydrogen PeroxideRe-evaluation of Some Organic Chemicals, Hydrazine and Hydrogen Peroxide
Your browsing activity is empty.
Activity recording is turned off.
See more...