NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.
This publication is provided for historical reference only and the information may be out of date.
Scope
NCBI’s PubChem BioAssay database (1-5) (http://pubchem.ncbi.nlm.nih.gov) is a public repository for archiving biological tests of small molecules and siRNA reagents. Small molecule bioactivity data contained in the BioAssay database consist of information generated through high-throughput screening experiments, medicinal chemistry studies, chemical biology research, as well as literature curation. In addition, the BioAssay database contains data from RNAi screens against targeted genes or complete genomes aiming to identify critical genes responsible for a biological process or disease condition. BioAssay data continue to grow rapidly and are integrated with the rest of the NCBI resources, making PubChem a widely used public information system for accelerating chemical biology research and drug development.
The mission of the PubChem resource is to deliver free and easy access to all deposited data, and to provide intuitive data analysis tools. The PubChem BioAssay database is organized as a set of relational databases deployed on Microsoft SQL servers. The infrastructure allows for seamlessly storing the submitted BioAssay records, tracking and versioning subsequent updates, and supporting data retrieval and analysis.
As a repository, PubChem constantly optimizes and develops its data submission system, answering many demands of both high and low volume depositors.
PubChem’s BioAssay data is integrated into the NCBI Entrez information retrieval system, thus making PubChem data searchable and accessible by Entrez queries. In addition, the PubChem information platform provides Web-based and programmatic tools for users to search, review, and download bioactivity data for a BioAssay record, a compound, a molecular target, or a publication. PubChem also provides a suite of integrated services enabling users to collect, compare, and analyze biological test results across multiple assay bioassay projects.
PubChem BioAssay Standard & Data Model
PubChem provides a flexible BioAssay data model (1,2) and database schema to accommodate bioactivity data produced by diverse experimental procedures. The data model continues to expand to support new types of information generated as experimental methodologies evolve.
An assay record is represented by a unique PubChem BioAssay accession, or AID. A BioAssay record is organized in two parts, the assay description and the assay results, and has links to the corresponding records of the substances that were tested by the assay, which are stored in the PubChem Substance database. Updates are tracked and a BioAssay record is versioned if any part of the record gets updated.
The assay description section includes an assay title, data source, assay description, experimental protocols, tested reagent category (e.g., small molecule vs siRNA), comments, assay targets, cross references to other databases at NCBI, and assay readout descriptions.
The assay result section includes the results for all tested substances. Results reported per substance can include regular assay readout, such as IC50 inhibition activity at a given test concentration. Per-substance assay data can also include annotations, including target description; comment on the individual biological test result; cross-links to other NCBI resources, such as Gene ID and PubMed ID (PMID); and URLs to the depositor’s website. Assay data are provided in a tabular format, with one tested substance per row and one assay test readout or annotation per column. A substance needs not have results reported for all defined test readouts. Multiple test result field definitions may be specified per assay, each with a unique test identifier (TID), name, description, data type, data unit, and annotation for cross-references. As a result, one can report replications of a specific readout as well as one or multiple series of dose-response data points. An example BioAssay record (Dose response biochemical screening assay for inhibitors of c-Jun N-Terminal Kinase 3 (JNK3)) can be accessed at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=1284.
Biological screening data submitted to PubChem is diverse and assay specific. As such, there are no specific requirements on the presence of particular test results or assay readouts; however, PubChem requires a summary result for each tested substance or chemical sample. The summary result is two-fold: bioactivity outcome and bioactivity score. The “bioactivity outcome” partitions results and includes five categories: chemical probe, active, inactive, inconclusive, and unspecified. The “bioactivity score” facilitates the separation of highly active compounds from the inactive ones. Many biological assays employ a dose-response scheme, with a primary endpoint. PubChem requires that this key readout, denoted as an “active concentration summary,” has micro-molar units, and that the experimental concentrations for the corresponding dose-response readouts (referred to as “tested concentrations,” also in micro-molar units) be designated on the respective test result fields as an attribution. These specialized readouts, together with the summary results, allow PubChem users to classify and rank hits of a screening test. They also support cross links from the BioAssay record to PubChem compounds, and allow PubChem to provide tools to enable in-depth data analysis and comparison across multiple BioAssay results.
PubChem BioAssay tracks the screening stage of a high throughput screening (HTS) assay project, if multiple BioAssay records are submitted for the project. The stages of an HTS project include: “screening,” a primary high-throughput assay where the activity outcome is based on percentage inhibition from a single dose; “confirmatory,” a low-throughput assay where the activity outcome is based on a dose-response relationship with multiple tested concentrations; “summary,” an assay summarizing information from multiple BioAssay submissions for validated chemical probes or small molecule leads ; and “other,” assays that do not fit the previous categories.
Assay targets are important information that should also be included in BioAssay records when possible. The “classical” assay model allows for the specification of assay target, either a single molecule or a complex, for the entire assay record, along with descriptions for the target molecules including gene and taxonomy information. In this model, the bioactivity outcomes provided in the entire assay dataset are solely for the specific target or target complex; for example, to describe the biological effect of the small molecules on the functionality of one enzyme.
PubChem also supports the presentation and annotation of multiple highly-related bioactivity outcomes, such as a profiling assay against a panel of molecular targets, in a single assay. Such a panel-type PubChem BioAssay record can contain multiple test readouts and respective bioactivity outcome annotations for each individual target, or similarly for each individual cell line or species defined within the “panel.” Each such target, cell line or species is regarded as a “panel component” in the data model, which can have its own “bioactivity outcome” or “active concentration” designated. An example panel assay (Kinase Inhibitor Selectivity Profiling Assay) can be accessed at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=1433.
A third bioassayBioAssay data model serves a general purpose for assays with multiple and substance- specific targets, but is primarily designed to support the representation of gene targets and test results for siRNA screenings, where each tested siRNA is aimed to suppress a specific gene target by design. This model allows one to specify a specific target and the relevant information for each individually tested sample (such as a siRNA reagent). As examples, an RNAi screening bioassayBioAssay (RNAi Global Initiative pilot viability screen of human kinase and cell cycle genes) can be accessed at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=1622; and a small molecule screening bioassayBioAssay (Experimentally measured binding affinity data derived from PDB) at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=1811.
PubChem tracks cross-references specified in a BioAssay submission, such as links to corresponding data in PubMed, Taxonomy, Gene, OMIM, or 3D structure of the target. In addition, the BioAssay data model distinguishes primary PubMed citations (references that contain experimental information directly relevant to the BioAssay record and can therefore aid the users’ interpretation and utilization of the assay data) from other PubMed citations that refer to or discuss the assay in a more general way.
In addition to providing data fields that capture essential information describing a BioAssay record, the BioAssay data model provides a flexible “categorized comment” mechanism that allows depositors to provide additional types of descriptive information, which are not explicitly listed as allowable tags in the data specifications document (http://pubchem.ncbi.nlm.nih.gov/upload/html/tags_assay.html). For example, this mechanism allows depositors to provide information pertinent to a focused research area, to comply with recommendations on a data standard from a working group, or to meet the guidelines of data exchange and sharing as required by a research community. Such a semi-structured data model also allows PubChem to accommodate a greater diversity of information critical to multiple research communities. An example, a BioAssay containing categorized comments (A CPE Based HTS Assay for Antiviral Drug Screening Against Dengue Virus) can be accessed at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=540333.
Tracking BioAssay Update
All data fields in a BioAssay record can be updated except the data source and the RegID (the tracking identifier provided by the depositor for the assay). An update to textual description or annotation triggers a version change for the assay description section (referred as description version). An update to a substance result is tracked by increasing the “test result” version. Duplicate tests and revisions to an existing test are both considered as test result updates. An update that involves a change to a test result definition (such as data type) or the addition or removal of test result fields triggers a major version change for the BioAssay record. For the major update, all BioAssay test results must be restated by the data depositor upon such fundamental changes. The BioAssay accession number, i.e., AID, remains unchanged upon these three types of updates. Description revision number and test result version number are associated with and counted against the major version of a BioAssay record. Whenever a major version is incremented, the description revision and test result version are reset to “1.” Only the current version of a description and corresponding test results are shown in the PubChem display system and indexed in the Entrez system by default, although all revisions are archived, tracked, and retrievable.
PubChem BioAssay Data Specification
The hierarchical data in the PubChem BioAssay archive is encoded in the data structure ASN.1 notation. All information about a single assay can be contained in a single ASN.1 or equivalent XML data object. It provides separate tagged fields for each aspect of the assay as detailed in the available specification in ASN.1 and XML Schema formats, respectively:
ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem.asn
ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem.xsd
Assay Neighboring and Related BioAssays
BioAssay records in PubChem can be related to each other in multiple ways. Several types of BioAssay relationships are computed in an automated way by PubChem. “Related BioAssays by Activity Overlap” tracks Bioassay records that share one or more active compounds. It allows one to rapidly identify, and thereby avoid, promiscuous inhibitors, or to help discover more complex target-based relationships. “Related BioAssays by Protein Target Similarity” tracks BioAssay records that have the same protein targets, or related protein targets (based on sequence similarity). It allows one to group compounds tested against the same or related targets; to isolate chemical agents with distinct biological effects, such as agonists and antagonists; or to evaluate selectivity of tested compounds. “Related BioAssays by BioSystems via Protein (or Gene) Target” tracks BioAssay records targeting on common biological pathways. This relationship identifies associations between genomic scanning studies that employed RNAi, and small molecule screening discovery studies that employed gene and protein targets. It allows one to take the responsible genes identified in RNAi knockdown experiments and identify small molecule therapeutics suggested in the small molecule screening tests. “Related BioAssays by Same Publication” links together BioAssay records that are extracted from the same publication, hence allows one to relate the results for better interpretation as illustrated in the publication.
Independent of computed BioAssay neighboring, “Related BioAssays” may be specified by the assay depositor. Normally, these relationships are specified when further confirmatory or counter-screenings are performed, thus providing the means to gather all screening data produced by the same screening campaign or assay project. Typically, a “Summary” assay is defined within such a grouping; it provides an overview of how each assay is involved in the overall effort, recaps the findings, and links to the individual assays as cross-references. To better support decision making, PubChem now also clusters and links up BioAssay records submitted for the same assay projects based on such “pair-wise” cross-references. Additional BioAssay relationship may be derived in future time, such as based on disease-target associations.
Public Access, Search, and FTP site
Data from the PubChem BioAssay database can be accessed via Web tools, direct Entrez queries, the FTP site, BLAST service, as well as from other NCBI databases that have links to PubChem (for example, a PubMed record about a medicine may contain a link to the corresponding PubChem record for that medicine).
A BioAssay record can be accessed by accession (AID) through the BioAssay Summary service at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=myAID, where “myAID” is a valid numeric PubChem BioAssay accession (AID). This service provides access to, and allows one to download, all deposited assay information, such as assay description, protocol and assay data. (As example, AID 1284 (“Dose response biochemical screening assay for inhibitors of c-Jun N-Terminal Kinase 3 (JNK3)) can be accessed at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=1284.) The service also lists information about the assay target, including depositor-provided molecular information and annotations derived by PubChem about protein family classification, the corresponding gene, pathway, and homologous 3D structures. Furthermore, the BioAssay Summary service provides a central entry point to a set of data analysis tools for the bioactive compounds identified in the assay. These analysis tools can be accessed through the “BioActivity Summary,” “Structure-Activity Analysis,” and “Structure Clustering” links that appear in the “BioActive Compounds” section of the assay record. They allow one to cluster the scaffolds of the tested compounds, examine, and visualize SAR relationships, and evaluate target specificity or promiscuity properties of the tested compounds. In addition, the “Related BioAssays” section lists assays that may be related to the one under review, and links to further detailed summaries of the BioAssay relationship. Cross-references to other NCBI databases, such as PubMed, are listed under the “Links” section.
The BioAssay database is indexed in Entrez and can be directly queried by entering text into the search box found on the BioAssay home page (http://www.ncbi.nlm.nih.gov/pcassay/), on the PubChem home page (http://pubchem.ncbi.nlm.nih.gov), or at the top of many PubChem Web pages. Descriptive information content in the BioAssay database is indexed under multiple fields to facilitate general as well as specific searches for BioAssay records. A full list of indexed fields and filters are documented at the PubChem Help page (http://pubchem.ncbi.nlm.nih.gov/help.html#PubChem_Index). For assistance with the construction of complex text queries or performing a specific search, one may use the “Limits” and “Advanced” search pages at http://www.ncbi.nlm.nih.gov/pcassay/limits and http://www.ncbi.nlm.nih.gov/pcassay/advanced respectively. Search results in Entrez are presented in tabular format where each row provides a result summary including assay title, data source, cross references, and links to the corresponding BioAssay record and assay data pages.
The BioAssay databsae is cross-linked to a number of other databases in Entrez, such as the PubChem Substance and Compound databases, PubMed, Entrez Protein, and Entrez Gene, and more. This makes it possible to access BioAssay even if you start your search in another database, by following the links from the record(s) you retrieve to the associated BioAssays data. In addition, the NCBI BLAST service allows one to search sequences of BioAssay targets. If any of the proteins listed on a BLAST results page were used as targets of BioAssays, they are flagged on the BLAST results page and linked to the BioAssay records. Integrating molecular sequence information of BioAssay targets with the BLAST service provides an additional path for biologists to discover and utilize the screening results within PubChem.
PubChem BioAssay FTP (ftp://ftp.ncbi.nlm.nih.gov/pubchem/BioAssay) provides open access to deposited BioAssay records. PubChem updates the BioAssay FTP site with new and modified BioAssay records on a daily basis in an incremental way. One can check the time stamp for the new post or update, and check the nature and history of an update by referring to the “assay.ftpdump.history” file at the FTP site. In addition to depositor-provided BioAssay records, annotations derived by PubChem from automated computation of BioAssay relationships can also be downloaded at ftp://ftp.ncbi.nlm.nih.gov/pubchem/BioAssay/AssayNeighbors/.
PubChem allows one to download BioAssay records in ASN, XML, and “comma-separated values” (CSV) formats. The structure of the FTP site is organized according to the respective data formats as shown in Figure 1, e.g., ASN and XML sub-directories provide BioAssay records containing both assay description and data in ASN.1 and XML format, respectively. The CSV sub-directory provides CSV-formatted assay data and XML-formatted assay description. The “Concise” directory contains the XML/ASN/CSV sub-directories with the same structure, but provides only summary assay results including bioactivity outcome, score, and active concentration. Because of the large number of BioAssay records, bulk downloads from the FTP site are now assisted by the “zip” compression of multiple records per file with BioAssay AID ranges in the filenames, such as “0000001_0001000.zip.”
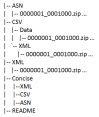
Figure 1.
PubChem BioAssay FTP directory structure.
Miscellaneous information, such as related BioAssay data, protein and gene target identifier lists, and a file containing a list of records (list of AIDs) that have been updated, etc., are also provided under various sub-directories at ftp://ftp.ncbi.nlm.nih.gov/pubchem/BioAssay.
BioAssay Tools
To make the vast bioactivity information easily accessible to the scientific community, PubChem provides a suite of integrated services enabling users to collect, compare and analyze biological test results, identify and validate drug targets, and evaluate chemical and RNAi probes.
BioAssay records can be accessed and downloaded at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=myAID, where “myAID” is a valid numeric PubChem BioAssay accession (AID). Plotting functions are provided for drawing dose-response curve and readout analysis through histogram and scatterplot. PubChem offers additional services for users to access and download summarized bioactivity information for a compound (http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?sid=mySID, http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=myCID), for a protein assay target (http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=myGI), and equivalently, for a gene (http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=myGeneID). Assay descriptions and data tables can also be retrieved and downloaded through programmatic interfaces using the PubChem Power User Gateway (PUG, http://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html), including PUG/SOAP (http://pubchem.ncbi.nlm.nih.gov//pug_soap/pug_soap_help.html) and PUG/REST (http://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html) facilities.
In addition, PubChem utilizes the summary results (e.g., active vs inactive) and specialized readouts (i.e., IC50) and provides Web-based tools for: 1) rapid data retrieval, analysis, integration, and comparison of biological screening results across multiple BioAssay records; 2) exploratory structure-activity analysis; 3) target selectivity examination; 4) reviewing related BioAssay records. These tools integrate chemical, target, literature, and biological activity information. They also support the navigation and in-depth data analysis that facilitates identification of bioactive compounds, study of biological profiling, and polypharmacology properties for drug or drug candidate molecules, and discovery of biological interesting targets contained within PubChem databases. A list of Web-based bioactivity analysis tools and their URLs are summarized in Table 1, which can also be accessed from the PubChem BioAssay home page at http://pubchem.ncbi.nlm.nih.gov/assay. The uses of these tools are described in detail in several publications (1-5).
Table 1.
A list of Web-based PubChem services for the BioAssay resource
BioAssay Submissions and Updates
PubChem Upload (http://pubchem.ncbi.nlm.nih.gov/upload/) supports the submission of chemical structures, BioAssay experimental results, annotations, drug targets, siRNAs, and more. The system provides an extensive set of wizards, inline help tips and guided tutorials to assist the submitter, based on their preference, to enter data and descriptive information by Web form or by file. PubChem Upload integrates convenient spreadsheet formats (CSV, Excel & OpenOffice) as well as XML-based data specifications to accommodate submitters of individual assays as well as institutional providers of data from large scale screening studies. A “Preview” facility displays incoming data in a mock record format to show how it will appear in PubChem before being released by the submitter. Such visual feedback to the submitter along with an automated suite of validation checks help insure data integrity and that everything appears as expected. Help documents and a tutorial provide an overview the PubChem Upload system and how it can be used:
The brief help document provides basic information about the PubChem Upload tool, including sample files for submitting substances and assays: http://pubchem.ncbi.nlm.nih.gov/upload/docs/upload_help.html
The complete help document includes the information provided in the brief document, plus technical details about the PubChem Upload tool and FTP submissions: http://pubchem.ncbi.nlm.nih.gov/upload/docs/upload_help_complete.html
The tutorial provides step-by-step examples of the procedure for submitting substances and/or bioassays to PubChem: http://pubchem.ncbi.nlm.nih.gov/upload/tutorial/
Summary
The PubChem BioAssay database is set up to serve as a public repository for bioactivity data of small molecules and RNAi. A streamlined information platform is provided at PubChem with a suite of tools allowing users to query the databases and analyze the retrieved BioAssay data. Integration with the Entrez system provides annotation services by linking small molecule modulators or effective RNAi reagents, as identified by screening experiments in the BioAssay database, to genomic and literature resources at NCBI. To meet the increasing demand from public users and from rapid growth of data volume and complexity, PubChem maintains and develops its service to the community as a public data repository by optimizing and expanding its BioAssay data model for supporting broader types of information, by developing infrastructure to ensure database scalability, by improving the deposition system to ease information exchange, and by enhancing search, retrieval, analysis, and download tools. PubChem welcomes the community to utilize the resource, provide feedback, and to further contribute data content to the repository.
References
- 1.
- Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T, Shoemaker BA, Gindulyte A, Bryant SH. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014 Jan;42(Database issue):D1075–82. Epub 2013 Nov 5. [PMC free article: PMC3965008] [PubMed: 24198245] [CrossRef]
- 2.
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, et al. PubChem's BioAssay Database. Nucleic Acids Res. 2012;40(Database issue):D400-12. Epub 2011/12/06. doi: 10.1093/nar/gkr1132 gkr1132 [pii]. [PMC free article: PMC3245056] [PubMed: 22140110] [CrossRef]
- 2.
- Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, et al. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38(Database issue):D255-66. Epub 2009/11/26. doi: gkp965 [pii] 10.1093/nar/gkp965. [PMC free article: PMC2808922] [PubMed: 19933261] [CrossRef]
- 3.
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37(Web Server issue):W623-33. [PMC free article: PMC2703903] [PubMed: 19498078]
- 4.
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: Integrated Platform of Small Molecules and Biological Activities. Annu Rep Comput Chem. 2008;4(Chapter 12):217-41.
- NCBI PubChem BioAssay Database - The NCBI HandbookNCBI PubChem BioAssay Database - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...
