NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.
This publication is provided for historical reference only and the information may be out of date.
Scope
The BioProject database provides an organizational framework to access information about research projects with links to data that have been or will be deposited into archival databases maintained at members of the International Nucleotide Sequence Database Consortium (INSDC, which comprises the DNA DataBank of Japan (DDBJ), the European Nucleotide Archive at European Molecular Biology Laboratory (ENA), and GenBank at the National Center for Biotechnology Information (NCBI)) (1,2,3).
BioProjects describe large-scale research efforts, ranging from genome and transcriptome sequencing efforts to epigenomic analyses to genome-wide association studies (GWAS) and variation analyses. Data are submitted to NCBI or other INSDC-associated databases citing the BioProject accession, thus providing navigation between the BioProject and its datasets. Consequently, the BioProject is a robust way to access data across multiple resources and multiple submission timepoints, e.g., when there are different types of data that had been submitted to multiple databases, or sequential submissions deposited over the course of a research project.
The definition of a set of related data, a “project,” is very flexible, so using different parameters allows the creation of a complex project and various distinct sub-projects. For example, BioProject records can be established for:
- Genome sequencing and assembly
- Metagenomes
- Transcriptome sequencing and expression
- Targeted locus sequencing
- Genetic or RH Maps
- Epigenomics and functional genomics
- Phenotype or Genotype
- Variation detection
The BioProject database encompasses taxonomic diversity, from humans and animals, to plants, to prokaryotes and metagenomes. BioProjects are created for initiatives that generate a very large volume of data, data from multiple members of a consortium or collaboration, or data being submitted to multiple archival databases. BioProject registration is required for some database submissions including dbVar, the Sequence Read Archive (DRA/ERA/SRA), and microbial and eukaryotic genome submissions to DDBJ/ENA/GenBank. However, small datasets that have one or a few sequences, like a single viral or organellar genome, are not in scope for BioProject.
The BioProject database defines two types of projects: 1) primary submission projects, as described above, are directly associated with submitted data and may be registered by submitters of that data using the NCBI submission portal; 2) umbrella projects, which reflect a higher-level organizational structure for larger initiatives or provide an additional level of data tracking. These projects are created by request. An umbrella project groups projects that are part of a single collaborative effort but represent distinct studies that differ in methodology, sample material, or research grant. Complex research efforts may be represented with more than one layer of umbrella project such that a highest-level umbrella project is linked to one or more sub-project umbrella projects which in turn are linked to one or more Primary submission projects that describe the data in more detail.
History
The BioProject resource became public in May 2011, replacing the older NCBI Genome Project database, which had been created to organize the genome sequences in GenBank and RefSeq (4). The BioProject database was created to meet the need for an organizational database for research efforts beyond just genome sequencing, such as transcriptome and gene expression, epigenomics, and variation studies. However, because a BioProject is defined by its multiple attributes, there is flexibility for additional types of projects in the future, beyond those that were included in 2011. The new BioProject database allows more flexible grouping of projects and can collect more data elements for each project, e.g., grant information and project relevance. Projects registered in the old Genome Project database were incorporated into BioProject, and a BioProject Accession was assigned in addition to the numerical ID that was previously assigned in Genome Project.
Data Model
Primary submission projects have attributes that describe the scope, methodology, and objectives of the project. The attributes are:
- Sample Scope indicates the sample purity and scope. The options are monoisolate, multiisolate, multi-species, environment, synthetic, and other.
- Material indicates the type of material isolated from the sample. The options include genome (for a genome or metagenome), purified chromosome, transcriptome, phenotype (phenotypic descriptive data), reagent (material studied was obtained by chemical reaction, precipitation), proteome (protein or peptide data).
- Capture indicates the scale, or type, of information that the study is designed to generate from the sample material. The options include whole (meaning that a specific subset was not used and which is the most common case), exome (capturing exon-specific data), and TargetedLocusLoci (specific loci such as a gene, genomic region, or barcode standard).
- Method is the general approach to generate the data. The options include sequence, array, and mass spectrometry.
- Objective is the project goals with respect to the type of data that will be generated and submitted to the data archives. Options include raw sequence reads (data to SRA); sequence, assembly and annotation (data to GenBank); expression (data to GenBank or GEO); variation (data to dbSNP or dbVAR); epigenetic markers (data to GEO or SRA) and phenotype (data to dbGaP).
The combination of attributes determines the project data type, (descriptive label), such as genome sequencing or epigenomics. Since multiple kinds of data, e.g., genome and transcriptome data, can be submitted with the same BioProject identifier, the project data type includes both values, Genome Sequencing and Transcriptome.
The BioProject also stores submitter and grant information, related publications, and links to external Web resources that are relevant for the project. Furthermore, the organism name, taxid, and infra-species identifier (strain, breed, cultivar, or isolate) of the Target of a project are currently stored in the BioProject database, and the organism name is refreshed daily by a lookup in the taxonomy database. However, by the end of 2014 the organism information will be maintained in the related BioSample database and only cached for display in the BioProject page.
The BioProject XML schema is presented on the FTP site, ftp://ftp.ncbi.nlm.nih.gov/bioproject/
Additional information about BioProject, including a glossary of terms, is available in the BioProject Help document.
Dataflow
Primary submission records may be created through the NCBI Submission Portal via several paths: (1) interactive Web portal, https://submit.ncbi.nlm.nih.gov/subs/bioproject/; (2) programmatic XML-based ui-less interface; (3) as part of data submission to some resources, such as GEO or dbGaP. In addition, RefSeq processes can create primary submission BioProjects.
Umbrella projects are created by NCBI staff at the request of submitters or funding agencies. Once an umbrella project exists, submitters link to the umbrella when creating a new primary BioProject. In addition, NCBI staff can create links between an umbrella project and pre-existing sub-projects at the submitter’s request.
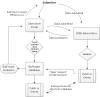
Error-free submissions are loaded into the database and assigned a BioProject accession, which has the format of five letters plus a series of digits, e.g., PRJNA31257. BioProjects are made public immediately unless a hold-until-published (HUP) date is requested. In that case, the BioProject is released on that HUP date or when the BioProject accession or the linked data is cited in a publication, or when data with that BioProject accession are released, whichever of those events occurs first. Public data in NCBI archives that include a BioProject accession trigger the creation of a reciprocal link in Entrez between the data record for that archive and the BioProject (Figure 1).
Creation of a BioProject is not sufficient for publication. The data that corresponds to that BioProject also needs to be submitted to the appropriate INSDC-associated database.
Public BioProjects are exchanged with the members of the INSDC nightly.
Access
In Entrez, BioProject records may be accessed by browsing, by query, by download, or by following a link from another NCBI database.
Browsing: From the BioProject home page users can navigate to the “By project attributes” hyperlink to browse through the database content by major organism groups, project type (umbrella projects vs. primary submissions), or project data type. The table includes links to the NCBI Taxonomy database where additional information about the organism may be available and to the BioProject record.
Query: Searches can be performed in BioProject like any other Entrez database, namely by searching for an organism name, text word, or BioProject accession (PRJNA31257), or using the Advanced Search page to build a query restricted by multiple fields. Search results can be filtered by Project Type, Project Attributes, Organism, or Metagenome Groups, or by the presence or absence of associated data in one of the data archives.
Here are some representative searches:
| Find BioProjects by… | Search text example(s) |
|---|---|
| A species name | Escherichia coli[organism] |
| Project data type | "metagenome"[Project Data Type] |
| Project data type and Taxonomic Class | "transcriptome"[Project Data Type] AND Insecta[organism] |
| Publication | "19643200"[PMID] |
| Submitter organization, consortium, or center | JGI[Submitter Organization] |
| Sample scope and material used | "scope environment"[Properties] AND "material transcriptome"[Properties] |
| A BioProject database identifier | PRJNA33823 or PRJNA33823[bioproject] or 33823[uid] or 33823[bioproject] |
Download: In addition to the Entrez Web interface and the BioProject browse page, users can download the entire database and the database .xsd schema from the FTP site, ftp://ftp.ncbi.nlm.nih.gov/bioproject/, or use Entrez Programming Utilities (E-utilities) to programmatically access public BioProject records.
Linking: BioProject records can be found by following links from archival databases when the data cites a BioProject accession. Links may be found in several databases including SRA, Assembly, BioSample, dbVar, Gene, Genome, GEO, and Nucleotide (which includes GenBank and RefSeq nucleotide sequences).
Report formats
Summary
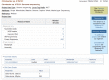
The Summary view provides a concise overview of the project and includes the BioProject name (which is often the organism name), title, Taxonomy, Project data type, Attributes, the project source, submitting organization, and the BioProject accession and ID (uid). The Project name or label is linked to the full report page, shown in Figure 2.
Full Report
The Full Report display for Primary submission projects, as shown in Figure 2, includes the project name and/or title, a text description of the project (when provided), the project data type and specific project attributes, a project data section with data links, citations relevant to the project, taxonomic lineage, information about the submitting group and project funding. Navigation tools are provided near the top of the report to facilitate navigation to NCBI’s taxonomically organized Genome resource, “up” to higher-level umbrella projects, or “across” to other BioProject records that are related by organism, or via a common umbrella project.
When the experimental data for a BioProject is submitted to archival databases, it contains the BioProject accession that links the data to the BioProject report page. The Project Data table in the report page presents data counts from those archival databases that have links to the BioProject. Genome sequencing BioProjects also have a table that reports the genome assembly’s accession number in the Assembly database, the BioSample accession, as well as the master accession number for whole genome sequencing (WGS) project, if relevant.
The page includes navigation tools to facilitate navigating to the related Genomes resource, which focuses on taxonomically organized genome sequencing projects, or to a linked umbrella project, or to “peer” projects that share a link to the same umbrella project or by shared taxonomy. If a genome assembly is represented by both an INSDC genome sequencing project, and a RefSeq genome project, then the correspondence between these projects is also indicated in the full report.
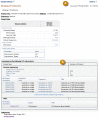
An umbrella project report page includes the relevant tabular reports listing the sub-projects that belong to that umbrella. The sub-projects may be 1) multiple primary submission projects of the same type, e.g., the HMP Reference Genome project PRJNA28331, 2) different kinds of primary submission bioprojects, e.g., PRJNA193500 (Figure 3), or 3) other umbrella projects, e.g., the HMP top-most project, PRJNA43021. The Data table of an umbrella project presents a sum of the data links for its grouped sub-projects, as seen for PRJNA193500 (Figure 3).
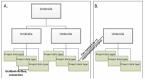
Some large initiatives are represented by more than one layer of umbrella projects (see Figure 4); for instance, a top-most level may identify the largest definition of the collaboration; a second level of umbrella projects identify the primary categories of data production; and finally a third layer represents the projects that actually generate the data that is submitted. The Human Microbiome project is an example of this type of complex hierarchy where the top-most project, PRJNA43021, represents the most inclusive definition of the initiative, and a secondary level (such as PRJNA28331) identifies a major sub-project to sequence multiple reference genomes each of which has a distinct project accession.
Related Tools
BioProjects may be registered with the submission portal at https://submit.ncbi.nlm.nih.gov/subs/bioproject/.
The submission portal is at https://submit.ncbi.nlm.nih.gov/subs/ and is designed to be a single place where submitters can register and deposit their data for multiple NCBI archives. As of November 2013, the submission portal is operational for BioProject, BioSample, WGS genomes, TSA transcriptomes, and the Genetic Testing Registry (GTR).
Other related resources include the BioSample, Assembly, and Genome databases. BioSample and BioProject are similar as they are both entry points for aggregating and retrieving data of a single research effort or sample from various NCBI databases.
The BioProject, Genome, and Assembly databases are interconnected and can be used to access and view genome assemblies different ways. Every prokaryotic and eukaryotic genome submission has BioProject, BioSample, Assembly, and GenBank accession numbers, so users can start in any of those resources and get to the others. The BioProject and BioSample databases allow users to find related data-sets, e.g., multiple bacterial strains from a single isolation location, or the transcriptome and genome from a particular sample. The Assembly accession is assigned to the entire genome and is used to unambiguously identify the set of sequences in a particular version of a genome assembly from a particular submitter. Finally, the Genome database displays all of the genome assemblies in INSDC and RefSeq, organized by organism.
References
- 1.
- Pruitt K, Clark K, Tatusova T, Mizrachi I. BioProject Help [Internet]. Bethesda (MD): National Center for Biotechnology Information; 2011 May. Available from: http://www
.ncbi.nlm.nih .gov/books/NBK54015. - 2.
- Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt K, Resenchuk S, Tatusova T, Yaschenko E, Ostell J. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2011;40(D1):D57–63. [PMC free article: PMC3245069] [PubMed: 22139929]
- 3.
- Nakamura Y, Cochrane G, Karsch-Mizrachi I., International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41(D1):D21–24. [PMC free article: PMC3531182] [PubMed: 23180798]
- 4.
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35(D1):D5–D12. [PMC free article: PMC1781113] [PubMed: 17170002]
- BioProject - The NCBI HandbookBioProject - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...