The content of this book is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 Unported license. To view the terms and conditions of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology [Internet]. 4th edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. doi: 10.1101/glycobiology.4e.12
Plasma membrane proteins are either peripheral proteins or integral membrane proteins. The latter include proteins that span the lipid bilayer once or several times, and a second class that are covalently attached to lipids. Proteins attached to glycosylphosphatidylinositol (GPI) via their carboxyl termini are generally found in the outer leaflet of the lipid bilayer facing the extracellular environment. The GPI membrane anchor may be conveniently thought of as an alternative to the single transmembrane domain of type-I integral membrane proteins. This chapter reviews the discovery, distribution, structure, biosynthesis, properties, and suggested functions of GPI anchors and related molecules, as well as their roles in diseases.
BACKGROUND AND DISCOVERY
The first tentative evidence for the existence of protein-phospholipid anchors appeared in 1963 with the finding that crude bacterial phospholipase C (PLC) selectively releases alkaline phosphatase from mammalian cells. Phosphatidylinositol (PI)-protein anchors were first postulated in the mid-1970s when highly purified bacterial PI-specific PLCs were observed to release proteins, such as alkaline phosphatase and 5′-nucleotidase, from mammalian plasma membranes. By 1985, these predictions were confirmed by compositional and structural data from studies on Torpedo acetylcholinesterase, human and bovine erythrocyte acetylcholinesterase, rat Thy-1, and the sleeping sickness parasite Trypanosoma brucei variant surface glycoprotein (VSG). The first complete GPI structures, which were for T. brucei VSG and rat Thy-1, were solved in 1988 (Chapter 1, Figure 1.3).
DIVERSITY OF PROTEINS WITH GPI ANCHORS
To date, hundreds of GPI-anchored proteins (GPI-APs) have been identified in many eukaryotes, ranging from protozoa and fungi to plants and humans (Online Appendix 12A). The range of described GPI-APs and the distribution of putative GPI biosynthesis genes suggests that (1) GPI anchors are almost ubiquitous among eukaryotes; (2) GPI-APs are functionally diverse and include hydrolytic enzymes, adhesion molecules, complement regulatory proteins, receptors, protozoan coat proteins, and prion proteins; and (3) in mammals, alternative messenger RNA (mRNA) splicing may lead to the expression of transmembrane and/or soluble and GPI-anchored forms of the same gene product. These variants may be developmentally regulated. For example, neural cell adhesion molecule (NCAM) exists in GPI-anchored and soluble forms when expressed in muscle and in GPI-anchored and two transmembrane forms when expressed in brain.
STRUCTURE OF GPI ANCHORS
Protein-Linked GPI Structures
The substructure Manα1-4GlcNα1-6myo-inositol-1-P-lipid is a universal hallmark of GPI anchors and related structures. All but one protein-linked GPI anchor share a larger common core structure (Figure 12.1; Online Appendix 12B, panels I–IV). The protein–carbohydrate association in GPI-APs is unique in that the reducing terminus of the GPI oligosaccharide is not attached to the protein but to the D-myo-inositol head group of a PI moiety by an α1-6 linkage. A distal, nonreducing mannose (Man) is attached to the protein via an ethanolamine phosphate (EtNP) bridge between its C-6 hydroxyl and the α-carboxyl group of the carboxy-terminal amino acid. GPIs are one of the rare instances in which GlcN is found without either an N-acetyl or N-sulfate (as in proteoglycans) moiety (Chapter 17).

FIGURE 12.1.
General structure of glycosylphosphatidylinositol (GPI) anchors attached to proteins. All characterized GPI anchors share a common core consisting of ethanolamine-PO4-6Manα1-2Manα1-6Manα1-4GlcNα1-6myo-inositol-1-PO4-lipid. (more...)
Beyond the common core, the structures of mature GPI anchors are quite diverse, depending on both the protein to which they are attached and the organism in which they are synthesized (Figure 12.1; Online Appendix 12B). Modifications to the core include additional EtNP and a wide variety of linear and branched glycosyl substituents of largely unknown function.
There is considerable variation in the PI moiety. Indeed, GPI is a rather loose term because, strictly speaking, PI refers specifically to D-myo-inositol-1-P-3(sn-1,2-diacylglycerol) (i.e., diacyl-PI), whereas many GPIs contain other types of inositolphospholipids, such as lysoacyl-PI, alkylacyl-PI, alkenylacyl-PI, and inositolphosphoceramide (Online Appendix 12B). Another variation is the attachment of an ester-linked fatty acid at the C-2 hydroxyl of the inositol residue, which makes the anchor inherently resistant to bacterial PI-PLCs. The available structural data suggest that (1) inositolphosphoceramide-based protein-linked GPIs are mainly found in “lower” eukaryotes, such as Saccharomyces cerevisiae, Aspergillus fumigatus, Aspergillus niger, Dictyostelium discoideum, and Trypanosoma cruzi; (2) the lipid structures of GPIs generally do not reflect those of the general cellular PI or inositolphosphoceramide pool; and (3) the lipid structures of some (e.g., trypanosome) GPI-APs are under developmental control.
The factors controlling the synthesis of a mature protein-linked GPI appear to be similar to those for other posttranslational modifications such as N- and O-glycosylation. Thus, primary control occurs at the cellular level, whereby the levels of specific biosynthetic and processing enzymes dictate the final repertoire of structures. Secondary control occurs at the level of the tertiary/quaternary structures of the GPI-APs, which affect accessibility to processing enzymes. Examples of primary control include (1) differences in GPI glycan side chains in human versus porcine membrane dipeptidase and brain versus thymocyte rat Thy-1 and (2) differences in glycan side chains and the lipid structure of T. brucei VSG in the bloodstream versus insect life-cycle stages of the parasite. An example of secondary control is the difference in VSG glycan side chains when VSGs with different carboxy-terminal sequences are expressed in the same trypanosome clone.
Non-Protein-Linked GPI Structures
In mammalian cells, some free GPIs (mature and biosynthetic intermediates) are found at the cell surface, although their functional significance remains unknown. On the other hand, several protozoa (particularly trypanosomatids) express high numbers (>107 copies per cell) of free GPIs on their cell surface as metabolic end products. These include the so-called glycoinositol phospholipids (GIPLs) and lipophosphoglycans (LPGs) of Leishmania. Some protozoan (type-1) GIPLs conform to the Manα1-6Manα1-4GlcNα1-6PI sequence common to protein-linked GPIs, whereas others contain a (type-2) Manα1-3Manα1-4GlcNα1-6PI motif, and still others possess hybrid structures containing the branched motif (Manα1-6)Manα1-3Manα1-4GlcNα1-6PI. The only GPI-linked fungal polysaccharide reported so far is A. fumigatus galactomannan-Manα1–2Manα1–2Manα1–6Manα1–4GlcN-inositolphosphoceramide, in which the glycan is attached directly to the anchor without the bridging EtNP (see Online Appendix 12B, panel II).
THE CHEMISTRY OF GPI ANCHORS
The GPI lends itself to selective cleavage by several chemical and enzymatic reagents (Online Appendix 12C, panel I). These were originally used to determine GPI structures and are now applied to confirm the presence, and/or obtain partial structural information, of GPIs. A key reaction is nitrous acid deamination of the GlcN residue, which selectively cleaves the GlcN-inositol glycosidic bond, thereby liberating the PI moiety that can be isolated by solvent partition and analyzed by mass spectrometry. The free reducing terminus, generated on the GPI glycan in the form of 2,5-anhydromannose, can be reduced to [1-3H]2,5-anhydromannitol (AHM) by sodium borotritide to introduce a radiolabel, or can be attached to a fluorophore such as 2-aminobenzamide (2-AB) by reductive amination. Once labeled and dephosphorylated with aqueous HF, the GPI glycan can be conveniently sequenced using exoglycosidases. Partial structural information can also be obtained by tandem mass spectrometry of (i) GPI-peptides generated by proteolytic digestion of GPI-APs or (ii) GPI glycans released by aqueous HF dephosphorylation and permethylated before analysis. Other indirect methods for inferring the presence of GPI-APs are described in Online Appendix 12C, panel II.
GPI BIOSYNTHESIS AND TRAFFICKING
The biosynthesis of GPI anchors occurs in three stages: (1) preassembly of a GPI precursor in the endoplasmic reticulum (ER) membrane, (2) attachment of the GPI to a newly synthesized protein in the ER lumen with concomitant cleavage of a carboxy-terminal GPI-addition signal peptide, and (3) lipid remodeling and/or carbohydrate side-chain modifications in the ER and after transport to the Golgi.
Analysis of biosynthesis of the GPI was made possible by the development of a cell-free system in T. brucei. The sequence of events underlying GPI biosynthesis has also been studied in T. cruzi, Toxoplasma gondii, Plasmodium falciparum, Leishmania major, Paramecium spp., S. cerevisiae, Candida albicans, Cryptococcus neoformans, and mammalian cells. The emphasis on eukaryotic microbes reflects the abundance of GPI-APs in these organisms and the potential of GPI inhibition for chemotherapeutic intervention. This notion has been genetically validated in the bloodstream form of T. brucei, as well as in P. falciparum, S. cerevisiae, and C. albicans.
The essential events in GPI biosynthesis are highly conserved. There are, however, variations on the theme. The T. brucei, S. cerevisiae, and mammalian cell GPI pathways are used here to showcase these differences (Figure 12.2). In all cases, GPI biosynthesis begins with the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to PI to produce GlcNAc-PI, which is de-N-acetylated in the next step to produce GlcN-PI on the cytoplasmic face of the ER (Figure 12.3; Table 12.1). Notable differences between the T. brucei and yeast or mammalian GPI-biosynthetic pathways occur from GlcN-PI onwards. Inositol acylation of GlcN-PI (at the C-2 hydroxyl of the D-myo-inositol; labeled as GlcN-aPI), strictly follows the first Man addition in T. brucei, whereas these steps are temporally reversed in yeast and mammalian cells. In the yeast and mammalian pathways, inositol acylation and deacylation are discrete steps occurring only at the beginning and end of the pathway, respectively, whereas in T. brucei these reactions occur on multiple GPI intermediates. Furthermore, in some mammalian cells such as human erythroblasts, inositol-deacylation never occurs and the mature GPI protein retains three hydrocarbon chains (Online Appendix 12B, panel III).
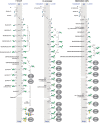
FIGURE 12.2.
Glycosylphosphatidylinositol (GPI)-biosynthetic pathways of Trypanosoma brucei, Saccharomyces cerevisiae, and mammals. These examples show that, despite the highly conserved core structure of GPI anchors, some diversity in GPI-biosynthesis exists. In (more...)
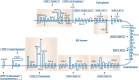
FIGURE 12.3.
Predicted topologies of the ER-resident components of glycosylphosphatidylinositol (GPI) biosynthesis in mammalian cells. Components within boxes belong to multisubunit complexes. The step numbers refer to those in Figure 12.2 and Table 12.1. The topologies (more...)
TABLE 12.1.
Components of the core mammalian and Saccharomyces cerevisiae glycosylphosphatidylinositol (GPI)-biosynthetic machinery
Fatty-acid remodeling in GPI of bloodstream from T. brucei occurs at the end of the pathway, but before transfer to protein, and involves exchanging the sn-2 fatty acids (a mixture of C18–C22 species) and the sn-1 fatty acid (C18:0) exclusively for C14:0 myristate. In yeast, this occurs in the ER, after transfer to proteins, and involves two distinct but consecutive processes. First, the unsaturated sn-2 fatty acid (C18:1) is exchanged for a C26:0 chain. Next, the diacylglycerol is exchanged for ceramide on many, but not all, GPI-APs. Lipid remodeling in mammalian cells is more complex. Many protein-linked GPIs contain sn-1-alkyl-2-acyl-PI with two saturated fatty chains, whereas the major cellular PI is sn-1-stearoyl-2-arachidonoyl-PI (i.e., with C18:0 and C20:4 fatty acids and few, if any, alkyl or alkenyl species). Two processes are involved in these structural changes. First, remodeling from the diacyl-PI to diradyl-PI (mixtures of 1-alkyl-2-acyl-PI and diacyl-PI), having unsaturated fatty acid at the sn-2-position, occurs early in the pathway, within the ER, to produce GlcN-aPI with the remodeled lipid tail. The reaction responsible for this is not known, but alkyl phospholipids synthesized in the peroxisome serve as the alkyl donors. Second, fatty-acid remodeling, involving exchange of the unsaturated sn-2 fatty acid for a saturated fatty acid, mainly stearate (C18:0), occurs after the GPI-APs are transported to the Golgi.
The GPI pathway genes were identified principally by expression cloning using GPI-deficient mutants of mammalian cells and temperature-sensitive yeast mutants. More recently, epitope tagging/pull-down/proteomic approaches have been used to identify GPI pathway–associated components. Some basic details of the known mammalian and yeast components and their topologies in the ER membrane are described in Table 12.1. Predicted topologies for the mammalian enzymes are shown in Figure 12.3.
The GPI precursor is transferred to proteins via a multisubunit transamidase complex. The reaction involves two complex substrates, the preassembled GPI precursor and the carboxyl terminus of a partially folded nascent protein (Figure 12.4). The carboxy-terminal GPI-addition signal peptide (GPIsp) contains three domains: (1) three relatively small amino acids (Ala, Asn, Asp, Cys, Gly, or Ser) located at ω, ω+1, and ω+2, where ω is the amino acid attached to the GPI anchor and ω+1 and ω+2 are the first two residues of the cleaved peptide; (2) a relatively polar domain of typically five to 10 residues; and (3) a hydrophobic domain typically comprising 15–20 hydrophobic amino acids. These GPIsp sequences have no strict consensus, but are easily identified by eye and by automated algorithms. The final hydrophobic stretch of amino acids often resembles a transmembrane domain, but the absence of positively charged and polar residues immediately downstream makes a GPIsp easy to spot. Like N-glycosylation sequons, a GPIsp will only be functional if the protein is translocated into the ER. Hence, all GPI-APs are synthesized with an amino-terminal ER localization signal peptide as well.
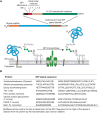
FIGURE 12.4.
(A) Features of glycosylphosphatidylinositol (GPI)-anchored proteins and their processing by GPI transamidase. GPI-anchored proteins have an amino-terminal signal peptide for translocation into the ER and a carboxy-terminal GPI-addition signal peptide (more...)
GPIs present microheterogeneity in their glycan side-chains depending on the species, cell types, and proteins (Online Appendix 12B, panels I–IV). A small number of enzymes involved in these reactions in mammals, yeast and T. brucei are known (Online Appendix 12B, panel V). Yeast and some mammalian GPIs possess a fourth α1-2Man (Man-4) residue added by Smp3/PIG-Z during GPI precursor assembly in the ER. Yeast GPI-APs have a fifth αMan (Man-5) residue added in the Golgi. A proportion of mammalian GPI-APs contain Man-1 modified by a β1-4N-acetylgalactosamine (GalNAc), and which may be further modified with a β1-4galactose (Gal), in the Golgi. An α2-3 sialic acid (Sia) could be added to the Gal. In T. brucei bloodstream forms, GPI-APs present glycan side-chains consisting of Gal, whereas the procyclic GPIs carry sialylated poly-N-acetyllactosamine and poly-lacto-N-biose structures.
The transport of GPI-APs from the ER to the Golgi is mediated by coat protein II (COPII)-coated transport vesicles. Their packaging into the vesicles requires a transmembrane cargo receptor, a complex of four p24 family proteins, that links luminally oriented GPI to COPII components on the cytoplasmic side of the ER membrane.
MEMBRANE PROPERTIES OF GPI-APs
GPI-APs with two long hydrocarbon chains (i.e., those containing diacylglycerol, alkylacylglycerol, alkenylacylglycerol, or ceramide) are stably associated with the lipid bilayer. It follows that inositol-acylated GPI proteins with three fatty-acid chains should be more stably associated. On the other hand, Leishmania LPG, with a single C24:0 alkyl chain, has a half-life of only minutes at the cell surface and is secreted intact into the medium.
The thermodynamics of bilayer interactions also depend on the length and degree of saturation of the fatty-acid chains. In this regard, the saturated nature of most (but not all) mammalian GPI anchors (Online Appendix 12B, panels III and IV) is thought to explain why GPI-APs associate with “lipid rafts.” The current model for mammalian lipid rafts is that of transient liquid-ordered nanoclusters of membrane components. These are dependent on dynamic cortical actin asters that, through adaptor proteins, cluster phosphatidylserine (PS) on the inner face of the plasma membrane bilayer. This coupling effect occurs because the long, saturated lipid chains from inner leaflet PS overlap and interact with those of GPI-APs and glycosphingolipids in the middle of the bilayer, thereby generating functional membrane domains. A compelling aspect of this model is that, although dynamic cortical actin can organize molecules across the leaflet into nanoclusters, the clustering of GPI-APs or glycosphingolipids at the outer leaflet can also, conversely, organize the inner leaflet and recruit molecules that may favor liquid-ordered domains. This provides a possible explanation for the perplexing, yet well-characterized, ability of GPI-APs to transduce signals across the plasma membrane.
There are many examples of transmembrane signaling via the cross-linking of GPI-APs with antibodies and clustering with a second antibody on various cells, particularly leukocytes. Cellular responses include an increase in intracellular Ca++, tyrosine phosphorylation, proliferation, cytokine induction, and oxidative burst. These signaling events are dependent on the presence of a GPI anchor and might be mostly caused by the induction and coalescence of lipid raft nanoclusters, although their participation in ligand binding in conjunction with signal receptors (such as receptor-like-kinases in Arabidopsis) to transduce extracellular signals cannot be ruled out. GPI-APs involved in transmembrane signaling, such as the glial-cell-(line-)derived neurotrophic factor receptor-α (GDNFR-α), need to be associated with transmembrane β coreceptors to transmit their signals. Similarly, GPI-anchored CD14 (the LPS/LPS-binding protein receptor) requires to partner transmembrane Toll-like receptor-4, and can function equally well with a GPI anchor or with a spliced transmembrane domain.
GPI ANCHORS AS TOOLS IN CELL BIOLOGY
The replacement of carboxy-terminal transmembrane domains of type-I integral membrane proteins by GPIsps allows their expression as GPI-APs on the plasma membrane of transfected mammalian cells. This offers a useful method for producing soluble forms of membrane proteins. For example, the T-cell receptor was rendered soluble after it was expressed in the GPI-anchored form (by the action of bacterial PI-PLC), and not by simply expressing the transmembrane-deleted domain. In addition, purified GPI-APs can be used to coat hydrophobic surface plasmon resonance chips, thus providing a convenient method for orienting and presenting proteins for binding studies. It is clear that purified GPI-APs will spontaneously insert into lipid bilayers. The physiological significance of direct GPI-protein exchange between membranes is still uncertain, particularly because all mammals express potent GPI-PLD activity in serum that can remove the lipid (phosphatidic acid) component of the anchor and, thus, prevent GPI-protein re-insertion. However, the re-insertion property of GPI-APs has been exploited experimentally to “paint” exogenous proteins onto cell surfaces.
BIOLOGICAL FUNCTIONS OF GPI ANCHORS
GPI anchors are essential for life in some, but not all, eukaryotic microbes. In S. cerevisiae, and probably most fungi, the presence of a GPI anchor targets certain mannoproteins for covalent incorporation into the β-glucan layer of the cell wall via a transglycosylation reaction, whereby Man-1 within the GPI-anchor core is transferred to the β-glucan polymer, probably by the actions of Dfg5 and Dcw1. Defects in cell wall biosynthesis are detrimental to yeast and this may be one of the reasons why GPI biosynthesis is essential for this organism. Certain unique phenotypes of C. albicans GPI biosynthetic mutants are strictly associated with a single gene defect, suggestive of highly specific cross-talk of the related GPI biosynthetic step with other cellular pathways. GPI biosynthesis is essential for the bloodstream form of T. brucei, even in tissue culture. This may be due to nutritional stress, given that this parasite uses an essential GPI-anchored transferrin receptor. On the other hand, surprisingly, GPI biosynthesis and/or transfer to protein are not essential for the insect-dwelling forms of T. brucei or L. major. In Arabidopsis thaliana, GPI biosynthesis is required for cell wall synthesis, morphogenesis, and pollen tube development. The availability of GPI-deficient mammalian cell lines shows that GPI-APs are not essential at the cellular level. However, mouse knockouts and tissue-specific conditional knockouts of the PIGA gene clearly show that GPI-APs are essential for early embryo and tissue development. GPI anchors impart to their attached proteins the ability to be shed from the cell surface in soluble form through the action of cellular or serum GPI-cleaving enzymes. Mammalian sperms acquire the ability to fuse with oocytes after GPI-anchored TEX101 and LY6K are released by a sperm-associated GPI-cleaving enzyme tACE (testis form angiotensin converting enzyme). The cleavage of GPI-APs is required for maturation of ADAM3, a metalloprotease on the sperm surface. Certain proliferating motor neurons initiate differentiation after the GPI-anchored proteinase inhibitor, RECK, is released by the GPI-cleaving enzyme, GDE2. This relieves the inhibition of ADAM10 metalloproteinase, allowing it to degrade the Notch-ligand to terminate Notch signaling. Thus, the cell switches from proliferation to differentiation. GPI-anchored CRIPTO acts as a coreceptor for Nodal signaling. Shedding of CRIPTO by a GPI-phospholipase A2, PGAP6, followed by a PLD-like enzyme, regulates Nodal signaling. In “lower” eukaryotes, GPI anchors may be useful for assembling particularly dense cell-surface protein coats, such as the VSG coat of T. brucei. In this case, each parasite expresses five million VSG dimers on the cell surface to protect it against complement-mediated lysis. If each VSG monomer had a single transmembrane domain instead of a GPI anchor, there would be little room for other integral membrane proteins such as hexose and nucleoside transporters. Generally, GPI-APs do recycle through intracellular compartments but, compared to typical transmembrane proteins, they reside in higher proportions on the cell surface and have longer half-lives. There are several examples of the exchange of GPI-APs from one cell surface to another. Some GPI-APs are incorporated into exosomes, suggesting the possibility of exosome-mediated cell-to-cell transfer. Sperm acquires some GPI-APs such as CD52, from epididymis, most likely mediated by exosomes.
GPI ANCHORS AND DISEASE
Paroxysmal nocturnal hemoglobinuria (PNH) is a human disease in which patients suffer from hemolytic anemia. The condition arises from loss of expression of several GPI-APs that protect the blood cells from lysis by the complement system (e.g., decay accelerating factor and CD59). The defect in PNH cells is a somatic mutation in the X-linked PIGA gene and appears to occur in a bone marrow stem cell. Unlike other enzymes in the pathway, which are encoded by autosomal genes, PNH caused by PIGA mutations is thought to arise at a higher frequency because of X inactivation. In both male and female stem cells, somatic mutation in the one active allele of PIGA results in complete loss of GlcNAc-T function (Chapter 46). Atypical PNH caused by PIGT or PIGB gene mutations has been reported; the patients show autoinflammatory features such as aseptic meningitis, in addition to typical PNH symptoms.
Inherited GPI deficiencies (IGDs) are caused by germline mutations in genes involved in GPI biosynthesis, protein transfer, and remodeling. Because complete GPI deficiency causes embryonic lethality, mutations in IGDs are hypomorphic, causing partial deficiency. Mutations in genes involved in GPI remodeling, such as in PGAP1, can be null and cause GPI-APs with abnormal structure. Patients with IGDs caused by mutations in 23 genes of the GPI-biosynthetic pathway have been reported (Table 12.1). Most of these mutations were identified by whole exome sequencing of patients’ cells. Major symptoms of IGDs are neurological problems, such as developmental delay/intellectual disability, seizures, cerebral and/or cerebellar atrophy, hearing loss, and visual impairment. Other symptoms include hyperphosphatasia; brachytelepharangy; abnormal facial features such as hypertelorism and tented mouth; cleft palate; anorectal, renal, and heart anomalies; and Hirschsprung disease (Chapter 45).
GPI biosynthesis and transfer to proteins are essential for yeast, for pathogenic fungi, and for the African sleeping sickness parasite T. brucei, as mentioned above. Several key surface molecules of the apicomplexan parasites, P. falciparum (malaria), Toxoplasma, and Cryptosporidium are GPI-anchored, and it is thought that the GPI pathway is likely to be essential in these pathogens. There is evidence that some parasite GPI anchors also play a direct role in modulating the host immune response to infection. Hence, pathogen-specific GPI pathway inhibitors are being actively sought as potential drugs (Online Appendix 12D). Indeed, Gwt1 inhibitors are currently being optimized as leads for antimalarial drugs. Gwt1 is also viewed as an important antifungal drug target. Of particular interest in this context is fosmanogepix, which is presently in phase-II clinical trials. Fungal GPI-APs shield cell wall β-(1,3)-glucans from detection by macrophages. Thus, inhibitors of GPI biosynthesis should support better clearance of the pathogen by the host immune system.
Like other glycoconjugates, GPI-APs can be exploited by pathogens. For example, the GPI anchors themselves are receptors for hemolytic pore-forming toxins, such as aerolysin from Aeromonas hydrophilia, which causes gastroenteritis, deep wound infections, and septicemia in humans. In addition, the GPI-AP CD55/DAF is the principal cell-surface ligand for enterovirus and several echoviruses. Finally, the conformational changes that the endogenous prion protein undergoes to become the aberrant spongiform-encephalopathy (“mad cow disease” or scrapie in sheep)-causing form may be associated with a clathrin-independent endocytic pathway that is followed by this GPI-anchored protein in neurons.
ACKNOWLEDGMENTS
The authors appreciate helpful comments and suggestions from Satyajit Mayor, Hiroshi Nakato, and Jerry Eichler.
FURTHER READING
- Ferguson MA, Williams AF. 1988. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu Rev Biochem 57: 285–320. doi:10.1146/annurev.bi.57.070188.001441 [PubMed: 3052274] [CrossRef]
- Ferguson MA, Homans SW, Dwek RA, Rademacher TW. 1988. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239: 753–759. doi:10.1126/science.3340856 [PubMed: 3340856] [CrossRef]
- Guha-Niyogi A, Sullivan DR, Turco SJ. 2001. Glycoconjugate structures of parasitic protozoa. Glycobiology 11: p45R–p59R. doi:10.1093/glycob/11.4.45r [PubMed: 11358874] [CrossRef]
- de Macedo CS, Shams-Eldin H, Smith TK, Schwarz RT, Azzouz N. 2003. Inhibitors of glycosylphosphatidylinositol anchor biosynthesis. Biochimie 85: 465–472. doi:10.1016/s0300-9084(03)00065-8 [PubMed: 12770785] [CrossRef]
- Maeda Y, Ashida H, Kinoshita T. 2006. CHO glycosylation mutants: GPI anchor. Methods Enzymol 416: 182–205. doi:10.1016/s0076-6879(06)16012-7 [PubMed: 17113867] [CrossRef]
- Levental I, Grzybek M, Simons K. 2010. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 49: 6305–6316. doi:10.1021/bi100882y [PubMed: 20583817] [CrossRef]
- Nikolaev AV, Al-Maharik N. 2011. Synthetic glycosylphosphatidylinositol (GPI) anchors: how these complex molecules have been made. Nat Prod Rep 28: 970–1020. doi:10.1039/c0np00064g [PMC free article: PMC5496678] [PubMed: 21448495] [CrossRef]
- Tsai YH, Liu X, Seeberger PH. 2012. Chemical biology of glycosylphosphatidylinositol anchors. Angew Chem Int Ed Engl 51: 11438–11456. doi:10.1002/anie.201203912 [PubMed: 23086912] [CrossRef]
- Guo Z. 2013. Synthetic studies of glycosylphosphatidylinositol (GPI) anchors and GPI-anchored peptides, glycopeptides, and proteins. Curr Org Synth 10: 366–383. doi:10.2174/1570179411310030003 [PMC free article: PMC4063365] [PubMed: 24955081] [CrossRef]
- Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, Suryawanshi S, Saikam V, Sawant SD, Panda A, et al. 2015. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 161: 581–594. doi:10.1016/j.cell.2015.03.048 [PMC free article: PMC4651428] [PubMed: 25910209] [CrossRef]
- Kinoshita T, Fujita M. 2016. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J Lipid Res 57: 6–24. doi:10.1194/jlr.r063313 [PMC free article: PMC4689344] [PubMed: 26563290] [CrossRef]
- Muñiz M, Riezman H. 2016. Trafficking of glycosylphosphatidylinositol anchored proteins from the endoplasmic reticulum to the cell surface. J Lipid Res 57: 352–360. doi:10.1194/jlr.r062760 [PMC free article: PMC4767001] [PubMed: 26450970] [CrossRef]
- Komath SS, Singh SL, Pratyusha VA, Sah SK. 2018. Generating anchors only to lose them: the unusual story of glycosylphosphatidylinositol anchor biosynthesis and remodeling in yeast and fungi. IUBMB Life 70: 355–383. doi:10.1002/iub.1734 [PubMed: 29679465] [CrossRef]
- Kinoshita T. 2020. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol 10: 190290. doi:10.1098/rsob.190290 [PMC free article: PMC7125958] [PubMed: 32156170] [CrossRef]
- BACKGROUND AND DISCOVERY
- DIVERSITY OF PROTEINS WITH GPI ANCHORS
- STRUCTURE OF GPI ANCHORS
- THE CHEMISTRY OF GPI ANCHORS
- GPI BIOSYNTHESIS AND TRAFFICKING
- MEMBRANE PROPERTIES OF GPI-APs
- GPI ANCHORS AS TOOLS IN CELL BIOLOGY
- BIOLOGICAL FUNCTIONS OF GPI ANCHORS
- GPI ANCHORS AND DISEASE
- ACKNOWLEDGMENTS
- FURTHER READING
- Review Glycosylphosphatidylinositol Anchors.[Essentials of Glycobiology. 2015]Review Glycosylphosphatidylinositol Anchors.Ferguson MAJ, Hart GW, Kinoshita T. Essentials of Glycobiology. 2015
- Review Glycosylphosphatidylinositol Anchors.[Essentials of Glycobiology. 2009]Review Glycosylphosphatidylinositol Anchors.Ferguson MAJ, Kinoshita T, Hart GW. Essentials of Glycobiology. 2009
- Review The Glycosylphosphatidylinositol Anchor: A Linchpin for Cell Surface Versatility of Trypanosomatids.[Front Cell Dev Biol. 2021]Review The Glycosylphosphatidylinositol Anchor: A Linchpin for Cell Surface Versatility of Trypanosomatids.Borges AR, Link F, Engstler M, Jones NG. Front Cell Dev Biol. 2021; 9:720536. Epub 2021 Nov 1.
- Synthetic analogues of glycosylphosphatidylinositol-anchored proteins and their behavior in supported lipid bilayers.[J Am Chem Soc. 2007]Synthetic analogues of glycosylphosphatidylinositol-anchored proteins and their behavior in supported lipid bilayers.Paulick MG, Wise AR, Forstner MB, Groves JT, Bertozzi CR. J Am Chem Soc. 2007 Sep 19; 129(37):11543-50. Epub 2007 Aug 23.
- Review Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling.[J Lipid Res. 2016]Review Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling.Kinoshita T, Fujita M. J Lipid Res. 2016 Jan; 57(1):6-24. Epub 2015 Nov 12.
- Glycosylphosphatidylinositol Anchors - Essentials of GlycobiologyGlycosylphosphatidylinositol Anchors - Essentials of Glycobiology
- Proteoglycans and Sulfated Glycosaminoglycans - Essentials of GlycobiologyProteoglycans and Sulfated Glycosaminoglycans - Essentials of Glycobiology
Your browsing activity is empty.
Activity recording is turned off.
See more...
