NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology [Internet]. 3rd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015-2017. doi: 10.1101/glycobiology.3e.017

Essentials of Glycobiology [Internet]. 3rd edition.
Show detailsThis chapter focuses on the structure, biosynthesis, and general biology of proteoglycans. Topics include a description of the major families of proteoglycans, their characteristic polysaccharide chains (glycosaminoglycans), biosynthetic pathways, and general concepts about proteoglycan function. Proteoglycans, like other glycoconjugates, have many essential roles in biology.
HISTORICAL PERSPECTIVE
The study of proteoglycans dates back to the beginning of the 20th century with investigations of “chondromucoid” from cartilage and anticoagulant preparations from liver (heparin). From 1930 to 1960, great strides were made in analyzing the chemistry of the polysaccharides of these preparations (also known as “mucopolysaccharides”), yielding the structure of hyaluronan (Chapter 16), dermatan sulfate (DS), keratan sulfate (KS), different isomeric forms of chondroitin sulfate (CS), heparin, and heparan sulfate (HS). Together, these polysaccharides came to be known as glycosaminoglycans (sometimes abbreviated as GAGs) to indicate the presence of amino sugars and other sugars in a polymeric form. Subsequent studies provided insights into the linkage of the chains to core proteins. These structural studies paved the way for biosynthetic studies.
The 1970s marked a turning point in the field, when improved isolation and chromatographic procedures were developed for the purification and analysis of tissue proteoglycans and glycosaminoglycans. Density-gradient ultracentrifugation allowed separation of the large aggregating proteoglycans from cartilage, revealing a complex of proteoglycan, hyaluronan, and link protein. Also during this period, it was realized that the production of proteoglycans was a general property of animal cells and that proteoglycans and glycosaminoglycans were present on the cell surface, inside the cell, and in the extracellular matrix (ECM). This observation led to a rapid expansion of the field and the eventual appreciation of proteoglycan function in cell adhesion, signaling, and other biological activities (Chapter 38). Today, studies with somatic cell mutants (Chapter 49) as well as experiments using gene knockout and silencing techniques in a variety of model organisms, such as nematode worms (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), African clawed frogs (Xenopus laevis), zebrafish (Danio rerio), and mice (Mus musculus), are aimed at extending our understanding of the role of proteoglycans in development and physiology (Chapters 25–27) and human diseases (Chapters 41–47). The application of a variety of newly developed analytical tools, including mass spectrometry (Chapter 50) and glycan arrays (Chapter 48), are leading to a better understanding of proteoglycan structure and function.
PROTEOGLYCAN AND GLYCOSAMINOGLYCAN COMPOSITION
Proteoglycans consist of a core protein and one or more covalently attached glycosaminoglycan chains (Figure 17.1). Glycosaminoglycans are linear polysaccharides, whose disaccharide building blocks consist of an amino sugar (glucosamine that is N-acetylated or N-sulfated or N-acetylgalactosamine) and a uronic acid (glucuronic acid or iduronic acid) or galactose. Figure 17.2 depicts short segments of glycosaminoglycans and their characteristic features. Hyaluronan does not occur covalently linked to proteoglycans but instead interacts noncovalently with some proteoglycans via hyaluronan-binding motifs (Chapter 16). Generally, invertebrates produce the same types of glycosaminoglycans as vertebrates, except that hyaluronan is not present and the chondroitin chains tend to be nonsulfated. Most proteoglycans also contain N- and O-glycans typically found on glycoproteins (see Chapters 9 and 10). The glycosaminoglycan chains are much larger than these other types of glycans (e.g., a 20-kDa glycosaminoglycan chain contains approximately 80 sugar residues, whereas a typical biantennary N-glycan contains 10 to 12 residues). Keratan sulfate is a sulfated poly-N-acetyllactosamine chain present on a limited number of proteins as an N-linked or O-linked chain. The composition of the glycosaminoglycan chains, the structure of the protein cores, and the distribution of the proteoglycan all determine the biological activities associated with proteoglycans.
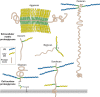
FIGURE 17.1.
Proteoglycans consist of a protein core (brown) and one or more covalently attached glycosaminoglycan chains (blue, HS ; yellow, chondroitin sulfate/dermatan sulfate). Membrane proteoglycans either span the plasma membrane (type I membrane proteins) or (more...)
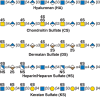
FIGURE 17.2.
Glycosaminoglycans consist of repeating disaccharide units composed of an N-acetylated or N-sulfated hexosamine and either a uronic acid (GlcA or IdoA) or galactose. Hyaluronan lacks sulfate groups, but the rest of the glycosaminoglycans contain sulfates (more...)
PROTEOGLYCANS ARE DIVERSE IN STRUCTURE AND FUNCTION
Virtually all mammalian cells produce proteoglycans and secrete them into the ECM, insert them into the plasma membrane, or store them in secretory granules. The ECM, an essential component of all multicellular animals, determines the physical characteristics of tissues and many of the biological properties of the cells embedded in it. The major components of the ECM are fibrillar proteins that provide tensile strength and elasticity (e.g., various collagens and elastins), adhesive glycoproteins (e.g., fibronectin, laminins, and tenascins), and proteoglycans that interact with other ECM components to promote ECM assembly, govern its physical properties, and serve as a reservoir of biologically active small proteins such as growth factors. A single cell type can express multiple proteoglycans. Vascular endothelial cells, for example, synthesize several different cell-surface proteoglycans, secretory granule proteoglycans, as well as several ECM proteoglycans.
Compared with the hundreds, perhaps thousands, of glycoproteins that carry N- and O-linked glycans, relatively few proteins have been identified that carry glycosaminoglycans (approximately 17 with heparan sulfate, approximately 20 with chondroitin/dermatan sulfate, and approximately eight with keratan sulfate). Nevertheless, tremendous structural variation of proteoglycans exists due to a number of factors. First, many proteoglycans can be substituted with one or two types of glycosaminoglycan chains; for example, glypicans contain heparan sulfate, whereas syndecan-1 contains both heparan sulfate and chondroitin sulfate chains. Some proteoglycans contain only one glycosaminoglycan chain (e.g., decorin), whereas others have more than 100 chains (e.g., aggrecan). Another source of variability lies in the stoichiometry of glycosaminoglycan chain substitution. For example, syndecan-1 has five attachment sites for glycosaminoglycans, but not all of the sites are used equally. Other proteoglycans can be “part-time”—that is, they may exist with or without a glycosaminoglycan chain or with only a truncated oligosaccharide. A given proteoglycan present in different cell types often shows differences in the number of glycosaminoglycan chains, their lengths, and the arrangement of sulfated residues along the chains. Thus, a preparation of any one proteoglycan (defined by its core protein) represents a diverse population of molecules, each potentially representing a unique structural entity. These characteristics, typical of all proteoglycans, create enormous diversity and potential biological variation in activity.
Mammalian Proteoglycans—Form and Function
The major classes of proteoglycans can be classified by their distribution, homologies, and function. Table 17.1 provides an overview of many of the known proteoglycans.
Table 17.1.
The vertebrate proteoglycans
The aggrecan family of ECM proteoglycans (also known as lecticans) consists of aggrecan, versican, brevican, and neurocan. In all four members, the protein moiety contains an amino-terminal domain capable of binding hyaluronan, a central region that contains covalently bound chondroitin sulfate chains, and a carboxy-terminal domain containing a C-type lectin domain (Chapter 34). Aggrecan is the best-studied member of this family, because it represents the major proteoglycan in cartilage where it forms a stable matrix capable of withstanding compressive forces by water desorption and resorption. Versican, which is produced predominantly by connective tissue cells, undergoes alternative splicing events that generate a family of proteins. Neurocan is expressed in the late embryonic central nervous system (CNS) and can inhibit neurite outgrowth. Brevican is expressed in the terminally differentiated CNS, particularly in perineuronal nets.
The small leucine-rich proteoglycans (SLRPs) contain leucine-rich repeats flanked by cysteines in their central domain. At least nine members of this family are known and some carry chondroitin sulfate, dermatan sulfate, or keratan sulfate chains. These proteoglycans help to stabilize and organize collagen fibers but have other roles in innate immunity and regulation of growth factor signaling.
The SLRPs and aggrecan family of proteoglycans appear to be unique to vertebrates. C. elegans and D. melanogaster express other proteoglycans, suggesting that the core proteins have undergone enormous diversification during evolution, presumably to accommodate different needs of the organism. In contrast, the biosynthetic machinery has been evolutionarily conserved, demonstrating conservation of function for the glycosaminoglycan chains.
Basement membranes are highly specialized thin layers of the ECM that lie flush against epithelial cells and surround muscle and fat cells. Major components are laminins, nidogens, and collagens, as well as three unrelated basement membrane proteoglycans, perlecan, agrin, and type XVIII collagen. These proteoglycans interact with other basement membrane components and cell-surface adhesion receptors, but may also be important reservoirs of heparan sulfate–binding growth factors.
The membrane-bound proteoglycans are diverse. The syndecan family consists of four members, each with a short hydrophobic domain that spans the membrane, linking the larger extracellular domain containing the glycosaminoglycan attachment sites to a smaller intracellular cytoplasmic domain. The syndecans are expressed in a tissue-specific manner and facilitate cellular interactions with a wide range of extracellular ligands, such as growth factors and matrix molecules. Because of their membrane-spanning properties, the syndecans can transmit signals from the extracellular environment to the intracellular cytoskeleton via their cytoplasmic tails. Syndecans are sensitive to proteolytic cleavage by matrix metalloproteases, resulting in shedding of the ectodomains bearing the glycosaminoglycan chains that retain potent biological activity (Chapter 38). C. elegans and D. melanogaster express only one syndecan (Chapter 26).
Glypicans carry only HS chains, which can bind a wide array of factors essential for development and morphogenesis. Six glypican family members exist in mammals, and only two are expressed in D. melanogaster and C. elegans. Each member of the glypican family of cell-surface proteoglycans has a glycosylphosphatidylinositol anchor attached at the carboxyl terminus, which embeds them in the outer leaflet of the plasma membrane (Chapter 12). The amino-terminal portion of the protein has multiple cysteine residues and a globular shape that distinguishes the glypicans from the syndecan ectodomains, which tend to be extended structures (Figure 17.1).
A number of other membrane proteoglycans are expressed on the surface of many different cell types including the widespread CD44, NG2 (also known as CSPG4), phosphacan (PTPζ), thrombomodulin, and invariant chain of the major histocompatibility complex (MHC) class II system. Serglycin is the major cytoplasmic secretory granule proteoglycan that is present in endothelial, endocrine, and hematopoietic cells. Depending on the species, it has a variable number of glycosaminoglycan attachment sites that can carry chondroitin sulfate or heparin chains. In fact, many proteoglycans show variation in the degree of substitution by GAG chains, giving rise in some cases to so-called “part-time” proteoglycans.
To a large extent, the biological functions of proteoglycans depend on the interaction of the glycosaminoglycan chains with different protein ligands. However, the protein core determines when and where expression takes place and then can interact with other components of the extracellular environment and the cytoskeleton. Table 17.2 lists examples of proteins known to interact with glycosaminoglycans. Proteins that bind to the sulfated glycosaminoglycan chains appear to have evolved by convergent evolution (i.e., they do not contain a specific fold present in all glycosaminoglycan-binding proteins, in contrast to other groups of glycan-binding proteins). These interactions have profound physiological effects and are discussed further in Chapter 38.
TABLE 17.2.
Examples of proteins that bind to sulfated glycosaminoglycans
LINKAGES OF GLYCOSAMINOGLYCANS TO PROTEINS
Different subtypes of sulfated glycosaminoglycans are attached to their core proteins by unique linkages. There are two types of keratan sulfate, distinguished by the nature of their linkage to protein (Figure 17.3). KS I, originally described in cornea, is found on an N-glycan linked to protein through an asparagine residue (Chapter 9). KS II (skeletal keratan sulfate) is found on an O-glycan core 2 structure and is thus linked through N-acetylgalactosamine (GalNAc) to serine or threonine (Chapter 10). The structural features in control of keratan sulfate substitution remain unclear, as the underlying poly-N-acetyllactosamine backbone can be found on many other glycoproteins. Notably, in humans and bovine, the large chondroitin sulfate proteoglycan found in cartilage (aggrecan) contains a segment of four to 23 hexapeptide repeats (E-E/L-P-F-P-S) where the keratan sulfate chains are located, whereas aggrecan in rats and other rodents lacks this motif and does not contain keratan sulfate.
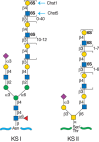
FIGURE 17.3.
Keratan sulfates contain a sulfated poly-N-acetyllactosamine chain-linked to either asparagine or serine/threonine residues. Chst1 and Chst5 add sulfate groups at the indicated positions. The actual order of the various sulfated and nonsulfated disaccharides (more...)
Two classes of glycosaminoglycan chains—chondroitin sulfate/dermatan sulfate and heparan sulfate/heparin—are linked to serine residues in core proteins by way of xylose (Figure 17.4). Xylosyltransferase initiates the process using UDP-xylose as donor. Two isoforms of the enzyme are known in vertebrates (Xylt1 and Xylt2), but only one isozyme exists in C. elegans and D. melanogaster. A glycine residue invariably lies to the carboxy-terminal side of the serine attachment site, but a perfect consensus sequence for xylosylation does not exist. At least two acidic amino acid residues are usually present, and they can be located on one or both sides of the serine, usually within a few residues. Several proteoglycans contain clustered glycosaminoglycan attachment sites, raising the possibility that xylosyltransferase could act in a processive manner. Xylosylation is an incomplete process in some proteoglycans, which may explain why proteoglycans with multiple potential attachment sites contain different numbers of chains in different cells.
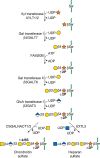
FIGURE 17.4.
The biosynthesis of chondroitin sulfate (left chain) and HS (right chain) is initiated by the formation of a linkage region tetrasaccharide. Addition of the first hexosamine commits the intermediate to either chondroitin sulfate (GalNAc) or heparan sulfate (more...)
After xylose addition, a linkage tetrasaccharide assembles by the transfer of two galactose residues catalyzed by unique members of the β4 galactosyl-, β3 galactosyl-, and β3 glucuronosyltransferase families of enzymes (Figure 17.4). This intermediate can undergo phosphorylation at the C-2 position of xylose and in the case of chondroitin sulfate, sulfation of the galactose residues. In general, phosphorylation and sulfation occur substoichiometrically, but phosphorylation may be transient. Phosphorylation occurs early in the assembly process and creates the preferred substrate for B4GALT7; a phosphatase removes the phosphate at a later stage of biosynthesis. Galactose sulfation is found in chondroitin sulfate, but its function remains unclear.
The linkage tetrasaccharide lies at a bifurcation in the biosynthetic pathway. Two types of reactions occur: addition of β4-linked N-acetylgalactosamine (GalNAc), which initiates chondroitin sulfate assembly, or addition of α4-linked N-acetylglucosamine (GlcNAc), which initiates heparan sulfate assembly (Figure 17.4). Genetic evidence from studies of C. elegans suggests that GalNAc addition during chondroitin assembly is mediated by the same enzyme that is involved in chain polymerization (SQV5), but biochemical evidence suggests that more than one enzyme may exist in vertebrates. In heparin/heparan sulfate formation, the addition of the first GlcNAc residue is catalyzed by an enzyme called EXTL3, which differs from the transferases involved in heparan polymerization (called EXT1 and EXT2). These enzymes are important control points because they ultimately regulate the type of glycosaminoglycan chain that will assemble. Control of the addition of β4GalNAc or α4GlcNAc appears to be manifested at the level of enzyme recognition of the polypeptide substrate.
GLYCOSAMINOGLYCAN BIOSYNTHESIS
Keratan Sulfate
Keratan sulfate chains contain a mixture of nonsulfated (Galβ4GlcNAcβ3), monosulfated (Galβ4GlcNAc6Sβ3), and disulfated (Gal6Sβ4GlcNAc6Sβ3) disaccharide units (Figure 17.2). The biosynthesis of the poly-N-acetyllactosamine backbone is described in Chapter 14. At least two classes of sulfotransferases, one or more GlcNAc 6-O-sulfotransferases (e.g., Chst6), and one Gal 6-O-sulfotransferase (Chst1) catalyze the sulfation reactions. These enzymes, like other sulfotransferases, use activated sulfate (PAPS [3′-phosphoadenyl-5′-phosphosulfate]) as a high-energy donor (Chapter 5). GlcNAc 6-O-sulfation occurs on the nonreducing terminal GlcNAc residue, as a prerequisite to further chain elongation, whereas sulfation of galactose residues takes place on nonreducing terminal and internal galactose residues, with a preference for galactose units adjacent to a sulfated GlcNAc. Sulfation of a nonreducing terminal galactose residue blocks further elongation of the chain, providing a potential mechanism for controlling chain length. The poly-N-acetyllactosamine chains of KS I are generally longer than those of KS II and may contain up to 50 disaccharide units (20–25 kDa). The relationship of enzymes involved in KS I and KS II sulfation is unclear. The chains can be fucosylated and sialylated as well (Chapter 14).
Chondroitin Sulfate
Vertebrate chondroitin sulfate consists of repeating sulfate-substituted GalNAcβ4GlcAβ3 disaccharide units polymerized into long chains (Figure 17.2). In contrast, invertebrates such as C. elegans and D. melanogaster make either nonsulfated or low sulfated chains, respectively. The assembly process for the backbone appears to be highly conserved, based on the presence of homologous genes for all of the reactions (Chapters 25 and 26). As described above, the assembly process is initiated by the transfer of GalNAcβ3 to the linkage tetrasaccharide (Figure 17.4). In both vertebrates and invertebrates, the polymerization step is catalyzed by one or more bifunctional enzymes (chondroitin synthases) that have both β3 glucuronosyltransferase and β4 N-acetylgalactosaminyltransferase activities. Vertebrates also express homologs that can transfer individual sugars to the chain. Chondroitin polymerization also requires the action of the chondroitin polymerizing factor (Chpf), a protein that lacks independent activity but collaborates with the polymerases to enhance the formation of polymers. Sulfation of chondroitin in vertebrates is a complex process, with multiple sulfotransferases involved in 4-O-sulfation and 6-O-sulfation of GalNAc residues (Figure 17.5).
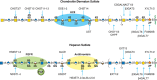
FIGURE 17.5.
Biosynthesis of chondroitin sulfate/dermatan sulfate involves the polymerization of GalNAc and GlcA units and a series of modification reactions including O-sulfation and epimerization of GlcA to IdoA. Heparan sulfate biosynthesis involves copolymerization (more...)
Additional enzymes exist for epimerization of glucuronic acid (GlcA) to iduronic acid (IdoA) in dermatan sulfate (Dse1-2), sulfation at the C-2-position of the uronic acids, and other patterns of sulfation found in unusual species of chondroitin (Table 17.3). The location of sulfate groups is easily assessed using bacterial chondroitinases (ABC, B, and ACII) that cleave the chains into disaccharides. Many chains are hybrid structures containing more than one type of chondroitin disaccharide unit. For example, dermatan sulfate is defined as having one or more IdoA-containing disaccharide units (chondroitin sulfate B) as well as GlcA-containing disaccharides (chondroitin sulfate A and C). Animal cells also degrade chondroitin sulfate in lysosomes using a series of exolytic activities (Chapter 44).
TABLE 17.3.
Types of chondroitin sulfates
Heparan Sulfate
Heparan sulfate assembles as a copolymer of GlcNAcα4GlcAβ4 (Figure 17.5), which then undergoes extensive modification reactions, catalyzed by at least four families of sulfotransferases and one epimerase. N-acetylglucosamine N-deacetylase/N-sulfotransferases (Ndst1-4) act on a subset of GlcNAc residues to generate N-sulfated glucosamine (GlcNSO3) units, many of which occur in clusters along the chain. Generally, the enzyme deacetylates GlcNAc and rapidly adds sulfate to the free amino group to form GlcNSO3, but a small number of glucosamine residues with unsubstituted amino groups may arise from incomplete N-sulfation. An epimerase (Glce), different from the one involved in dermatan sulfate synthesis, then acts on some GlcA residues, followed by 2-O-sulfation of some of the IdoA units (catalyzed by Hs2st). Some glucuronic units also undergo 2-O-sulfation by the same enzyme. The addition of 2-O-sulfate groups to GlcA or IdoA blocks the epimerization reaction. Next, 6-O-sulfotransferases (Hs6st1-3) add sulfate groups to selected glucosamine residues. Finally, certain subsequences of sulfated sugar residues and uronic acid epimers provide targets for 3-O-sulfotransferases (Hs3st1-6).
In contrast to chondroitin chains, which tend to have long tracts of fully modified disaccharides, the modification reactions in heparan sulfate biosynthesis occur in clusters along the chain, with regions devoid of sulfate separating the modified tracts. In general, the reactions proceed in the order indicated, but they often fail to go to completion, resulting in tremendous chemical heterogeneity. The disaccharide composition of the chains can be readily assessed using bacterial heparin lyases or chemical degradation methods (which are more useful for differentiating GlcA/IdoA) but direct sequencing of the chains has proven difficult because of their heterogeneity. New mass spectrometry methods are making significant inroads into sequencing of glycosaminoglycans (Chapter 50).
Regulation of Glycosaminoglycan Assembly
The specific arrangement of sulfated residues and uronic acid epimers in heparin/heparan sulfate and dermatan sulfate gives rise to binding sequences for ligands. The three examples shown in Figure 17.5 show minimal sequences that can interact with fibroblast growth factors (FGFs), antithrombin, and heparin cofactor II. More modified sequences can interact as well, and the binding of most FGFs is actually more sensitive to overall sulfation than to the specific position of the sulfate groups. Binding of glycosaminoglycans to proteins is described in greater detail in Chapter 38. A major question remains regarding how the enzymes and biosynthetic pathways are regulated to achieve tissue-specific expression of ligand-binding sequences.
During the last decade, most if not all of the enzymes involved in glycosaminoglycan synthesis have been purified and molecularly cloned from mammals and model organisms. Several important features have emerged from these studies, which may shed light on how different protein-binding sequences arise.
- Several of the enzymes appear to have dual catalytic activities. Thus, a single protein bearing two catalytic domains catalyzes N-deacetylation of GlcNAc residues and subsequent N-sulfation (Ndsts) in heparan sulfate formation. The same is true of the copolymerases, which transfer GlcNAc and GlcA (heparan sulfate) and GalNAc and GlcA (chondroitin sulfate) from the corresponding UDP sugars to the growing polymer. In contrast, the epimerases and O-sulfotransferase activities appear to be unique properties of independent enzymes.
- In several cases, multiple isozymes exist that can catalyze either a single or a pair of reactions. Thus, four N-deacetylase/N-sulfotransferases, three 6-O-sulfotransferases, and seven 3-O-sulfotransferases have been identified in heparan sulfate biosynthesis. Their tissue distribution varies and differences exist in substrate preference, which may cause differences in the pattern of sulfation. However, some overlap in expression and in substrate utilization occurs as well. Multiple isozymes of 4-O- and 6-O-sulfotransferases also can participate in chondroitin sulfate formation.
- The polymerization and polymer modification reactions probably colocalize in the same stacks of the Golgi complex. Thus, the enzymes may form supramolecular complexes that coordinate these reactions. The composition of these complexes may play a part in regulating the fine structure of the chains.
- In general, the composition of heparan sulfate, and likely chondroitin and dermatan sulfate, on a given proteoglycan varies more between cell types than that of heparan sulfate on different core proteins expressed in the same cell. This observation suggests that each cell type may express a unique array of enzymes and potential regulatory factors. The mechanisms behind the generation of apparently cell-specific glycosaminoglycan chains through regulated, yet partly stochastic modification reactions remain poorly understood.
- Recombinant enzymes and new synthetic schemes are increasingly used to generate defined glycosaminoglycan oligosaccharides, which can be used to probe ligand-binding affinities and specificities.
HEPARIN VERSUS HEPARAN SULFATE
Considerable confusion exists regarding the definition of heparin and heparan sulfate. Heparin is produced by a limited number of cells, notably connective tissue-type mast cells and bipotential glial progenitor cells, whereas heparan sulfate is made by virtually all types of cells. During biosynthesis, heparin undergoes more extensive sulfation and uronic acid epimerization, such that >80% of the GlcNAc residues are N-deacetylated and N-sulfated and >70% of the glucuronic acids undergo epimerization to IdoA. Heparin derived from porcine entrails and bovine lung is prepared commercially by selective precipitation and is sold by pharmaceutical companies as an anticoagulant because of its capacity to bind to antithrombin (Chapters 38 and 57). The active sequence is a pentasaccharide shown in Figure 17.5, which is now sold as a purely synthetic anticoagulant (Arixtra). Low-molecular-weight heparins are derived from commercial unfractionated heparin by chemical or enzymatic cleavage, depending on the brand. Selectively desulfated forms of heparin are also available commercially, some of which lack anticoagulant activity, but still retain other potentially useful properties (e.g., inhibition of inflammation and cell proliferation, and antimetastatic activity). Heparan sulfate also can contain anticoagulant activity, but typical preparations from cells or tissues are much less active than heparin. Care should be taken in extrapolating data obtained with heparin (e.g., binding to heparin-Sepharose) versus binding to heparan sulfate and heparan sulfate proteoglycans; binding to heparin can occur owing to the high charge content of the polysaccharide, whereas the same factor might bind to heparan sulfate with lower affinity or not at all. On the other hand, specific protein-binding motifs expressed in subspecies of heparan sulfate may occur also in heparin, although concealed by additional, redundant, sulfate residues.
PROTEOGLYCAN PROCESSING AND TURNOVER
Cells secrete matrix proteoglycans directly into the extracellular environment (e.g., members of the aggrecan family, the basement membrane proteoglycans, SLRPs, and serglycin). However, others are shed from the cell surface through proteolytic cleavage of the core protein through matrix metalloproteases (e.g., the syndecans). Cells also internalize a large fraction of cell-surface heparan sulfate proteoglycans by endocytosis. These internalized proteoglycans first encounter proteases and heparanase, an endo-β-glucuronidase, and the core protein and the heparan sulfate chains are cleaved endolytically. The resulting smaller heparan sulfate fragments eventually appear in the lysosome and undergo complete degradation by way of a series of exoglycosidases and sulfatases (Chapter 44). Chondroitin sulfate and dermatan sulfate proteoglycans follow a similar endocytic route. One of the human hyaluronidases (Hyal-4) has been found to be involved in endolytic degradation of chondroitin sulfate.
Cells secrete heparanase as well. Extracellular heparanase can cleave heparan sulfate chains at restricted sites, resulting in release of growth factors or chemokines immobilized on heparan sulfate proteoglycans at cell surfaces or in the ECM. In particular, invading cells secrete heparanase. Thus, heparanase may act with matrix metalloproteases to remodel the ECM.
A family of plasma membrane endosulfatases (Sulfs) can remove sulfate groups from internal 6-O-sulfated glucosamine residues in heparan sulfate. This postassembly processing of the chains at the cell surface results in altered response of cells to growth factors and morphogens. The mammalian genome contains other sulfatases of unknown function, raising the possibility that other postassembly processing reactions of glycosaminoglycans may occur.
ACKNOWLEDGMENTS
The authors appreciate helpful comments and suggestions from Andreas Geissner, Marissa Martinez, and Yuko Naito-Matsui.
FURTHER READING
- Yurchenco PD. 2011. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 3: a004911. [PMC free article: PMC3039528] [PubMed: 21421915]
- Lindahl U, Kjellén L. 2013. Pathophysiology of heparan sulphate: Many diseases, few drugs. J Intern Med 273: 555–571. [PubMed: 23432337]
- Thacker BE, Xu D, Lawrence R, Esko JD. 2013. Heparan sulfate 3-O-sulfation: A rare modification in search of a function. Matrix Biol 35: 60–72. [PMC free article: PMC4039620] [PubMed: 24361527]
- Vlodavsky I, Iozzo RV, Sanderson RD. 2013. Heparanase: Multiple functions in inflammation, diabetes and atherosclerosis. Matrix Biol 32: 220–222. [PubMed: 23499526]
- Filmus J, Capurro M. 2014. The role of glypicans in Hedgehog signaling. Matrix Biol 35: 248–252. [PubMed: 24412155]
- Wight TN, Kinsella MG, Evanko SP, Potter-Perigo S, Merrilees MJ. 2014. Versican and the regulation of cell phenotype in disease. Biochim Biophys Acta 1840: 2441–2451. [PMC free article: PMC4074575] [PubMed: 24401530]
- Xu D, Esko JD. 2014. Demystifying heparan sulfate–protein interactions. Annu Rev Biochem 83: 129–157. [PMC free article: PMC7851832] [PubMed: 24606135]
- Friand V, David G, Zimmermann P. 2015. Syntenin and syndecan in the biogenesis of exosomes. Biol Cell 107: 331–341. [PubMed: 26032692]
- Gallagher J. 2015. Fell-Muir Lecture: Heparan sulphate and the art of cell regulation: A polymer chain conducts the protein orchestra. Int J Exp Pathol 96: 203–231. [PMC free article: PMC4561558] [PubMed: 26173450]
- Hultgardh-Nilsson A, Boren J, Chakravarti S. 2015. The small leucine-rich repeat proteoglycans in tissue repair and atherosclerosis. J Intern Med 278: 447–461. [PMC free article: PMC4616156] [PubMed: 26477596]
- Mizumoto S, Yamada S, Sugahara K. 2015. Molecular interactions between chondroitin-dermatan sulfate and growth factors/receptors/matrix proteins. Curr Opin Struct Biol 34: 35–42. [PubMed: 26164146]
- Neill T, Schaefer L, Iozzo RV. 2015. Decoding the matrix: Instructive roles of proteoglycan receptors. Biochemistry 54: 4583–4598. [PMC free article: PMC4859759] [PubMed: 26177309]
- Review Proteoglycans and Sulfated Glycosaminoglycans.[Essentials of Glycobiology. 2022]Review Proteoglycans and Sulfated Glycosaminoglycans.Merry CLR, Lindahl U, Couchman J, Esko JD. Essentials of Glycobiology. 2022
- Review Proteoglycans and Sulfated Glycosaminoglycans.[Essentials of Glycobiology. 2009]Review Proteoglycans and Sulfated Glycosaminoglycans.Esko JD, Kimata K, Lindahl U. Essentials of Glycobiology. 2009
- Presence of unsulfated heparan chains on the heparan sulfate proteoglycan of human colon carcinoma cells. Implications for heparan sulfate proteoglycan biosynthesis.[J Biol Chem. 1989]Presence of unsulfated heparan chains on the heparan sulfate proteoglycan of human colon carcinoma cells. Implications for heparan sulfate proteoglycan biosynthesis.Iozzo RV. J Biol Chem. 1989 Feb 15; 264(5):2690-9.
- Differentially expressed patterns of glycosaminoglycan structure in heparan sulfate proteoglycans and free chains.[Eur J Biochem. 1993]Differentially expressed patterns of glycosaminoglycan structure in heparan sulfate proteoglycans and free chains.Hovingh P, Piepkorn M, Linker A. Eur J Biochem. 1993 Feb 1; 211(3):771-9.
- Isolation and characterization of proteoglycans synthesized by cultured mesangial cells.[J Biol Chem. 1990]Isolation and characterization of proteoglycans synthesized by cultured mesangial cells.Yaoita E, Oguri K, Okayama E, Kawasaki K, Kobayashi S, Kihara I, Okayama M. J Biol Chem. 1990 Jan 5; 265(1):522-31.
- Proteoglycans and Sulfated Glycosaminoglycans - Essentials of GlycobiologyProteoglycans and Sulfated Glycosaminoglycans - Essentials of Glycobiology
Your browsing activity is empty.
Activity recording is turned off.
See more...

