NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009.

Essentials of Glycobiology. 2nd edition.
Show detailsN-glycans are covalently attached to protein at asparagine (Asn) residues by an N-glycosidic bond. Five different N-glycan linkages have been reported, of which N-acetylglucosamine to asparagine (GlcNAcβ1-Asn) is the most common. This chapter describes only the GlcNAcβ1-Asn N-glycans, their general structure, the steps in their synthesis and processing, and the origins of their structural diversity. Terminal sugars that determine much of the diversity of N-glycans are referred to only briefly because they are described in detail in Chapter 13. Similarly, a discussion of glycosylation-mediated quality control of protein folding is reserved for Chapter 36, and the synthesis of the mannose-6-phosphate (Man-6-P) recognition determinant necessary for targeting lysosomal hydrolases to lysosomes is described in Chapter 30. Congenital disorders of glycosylation that arise from defects in N-glycan synthesis are discussed in Chapter 42.
DISCOVERY AND BACKGROUND
The GlcNAcβ1-Asn linkage was initially discovered by biochemical analyses of abundant glycoproteins found in serum, such as immunoglobulins. Importantly, early experiments on glycoprotein synthesis showed that not all asparagine residues can accept an N-glycan. The minimal amino acid sequence begins with asparagine followed by any amino acid except proline and it ends with serine or threonine (Asn-X-Ser/Thr). Thus, Asn-X-Ser/Thr “sequons” in a protein are candidates for receiving an N-glycan. A fascinating aspect of N-glycans is their complicated biosynthesis. Sensitive and rapid labeling techniques using [2-3H]-mannose revealed how these glycans are initially synthesized on a lipid-like molecule termed dolichol phosphate (Dol-P), followed by “en bloc” transfer of the entire glycan of 14 sugars to protein. This synthetic pathway is conserved in all of the metazoa, in plants, and in yeast. Bacteria use a related pathway to synthesize their cell wall. Other linkages to asparagine have been described, including glucose to asparagine in laminin of both mammals and Archaea, N-acetylgalactosamine (GalNAc) to asparagine in Archaea, and rhamnose to asparagine in bacteria. In a sweet corn glycoprotein, arginine is found in N-linkage to glucose.
Understanding these N-glycan pathways is important because N-glycans affect many properties of glycoproteins including their conformation, solubility, antigenicity, and recognition by glycan-binding proteins. N-glycans are used as tags by cell biologists to localize a glycoprotein or to follow its movement through the cell. Defects in N-glycan synthesis lead to a variety of human diseases.
MAJOR STRUCTURAL CLASSES AND NOMENCLATURE
All N-glycans share a common core sugar sequence, Manα1–6(Manα1–3)Manβ1–4GlcNAcβ1–4GlcNAcβ1-Asn-X-Ser/Thr, and are classified into three types: (1) oligomannose, in which only mannose residues are attached to the core; (2) complex, in which “antennae” initiated by N-acetylglucosaminyltransferases (GlcNAcTs) are attached to the core; and (3) hybrid, in which only mannose residues are attached to the Manα1–6 arm of the core and one or two antennae are on the Manα1–3 arm. An example of each N-glycan type is given in Figure 8.1.

FIGURE 8.1
Types of N-glycans. N-glycans added to protein at Asn-X-Ser/Thr sequons are
of three general types in a mature glycoprotein: oligomannose, complex, and
hybrid. Each N-glycan contains the common core
Man3GlcNAc2Asn. Symbol Key:

PREDICTING SITES OF N-GLYCOSYLATION
N-Glycans occur on many secreted and membrane-bound glycoproteins at Asn-X-Ser/Thr sequons. Analyses of protein sequence databases have revealed that about two thirds of the entries contain this consensus sequence. It is estimated that at least two thirds of those sequons are likely to be N-glycosylated. Occasionally, N-glycans are found at Asn-X-Cys, provided that the cysteine is in the reduced form. Although there have been several published reports of nucleocytoplasmic or cytoplasmic N-glycans, there exists no definitive structural evidence that N-glycans actually occur on cytoplasmic or nuclear proteins nor on the cytoplasmic portions of membrane proteins. The transfer of N-glycans to Asn-X-Ser/Thr sequons occurs on the lumenal side of the endoplasmic reticulum (ER) membrane while the protein moiety is being synthesized on ER-bound ribosomes and is translocating through the translocon in the ER membrane. Membrane glycoproteins remain anchored in the ER membrane with portion(s) exposed to the ER lumen, other portions embedded in the membrane, and yet other regions within the cytoplasm. Only domains that are accessible to the ER lumen will receive an N-glycan. Glycoproteins that lack a transmembrane domain also receive N-glycans cotranslationally, and ultimately they translocate completely into the lumen of the ER.
It is important to note that whereas the presence of the Asn-X-Ser/Thr sequon is necessary for the receipt of an N-glycan, transfer of the N-glycan to this sequon does not always occur, due to conformational or other constraints during glycoprotein folding. Also, the identity of “X” may reduce the efficiency of glycosylation, such as when “X” is acidic (aspartate or glutamate). Thus, when Asn-X-Ser/Thr sequons are present in a deduced amino acid sequence encoded by a cDNA, they are not identified categorically as N-glycan sites, but are referred to as potential N-glycan sites. Proof that an N-glycan is actually present at a potential site requires experimental evidence, as described later in this chapter.
ISOLATION, PURIFICATION, AND ANALYSIS
N-Glycans may be released from asparagine using a bacterial enzyme known as peptide-N-glycosidase F (N-glycanase F, PNGase F). This enzyme will remove oligomannose, hybrid, and complex N-glycans attached to asparagine. However, it will not remove N-glycans with certain modifications of the N-glycan core found so far only in slime molds, plants, insects, and parasites. Another bacterial enzyme termed PNGase A will remove these structures as well as all structures removed by PNGase F. Both enzymes are amidases that release N-glycans attached to the nitrogen of asparagine, thereby converting asparagine to aspartate. Therefore, sites of glycosylation can be deduced by amino acid sequence analysis performed before and after PNGase F treatment. Other bacterial enzymes cleave between the two core N-acetylglucosamine residues, leaving one N-acetylglucosamine attached to asparagine. These endoglycosidases are more specific in terms of the N-glycan structures they will cleave. Endoglycosidase H will release oligomannose and hybrid N-glycans, but not complex N-glycans. Endoglycosidase F will release simple biantennary N-glycans, but not oligomannose or hybrid N-glycans. N-Glycans may also be obtained free of protein by hydrazinolysis or by exhaustive digestion with a protease that removes all amino acids except for the asparagine. Released N-glycans may be purified by conventional ion-exchange and size-exclusion chromatography, high-pressure liquid chromotography (HPLC) methods, and affinity chromatography on glycan-binding proteins called lectins. Lectins for glycan analysis are usually obtained from plants and are described briefly in Chapter 1 and in detail in Chapters 29 and 45. Purification and analytical methods for obtaining and characterizing N-glycans are described in Chapter 47.
SYNTHESIS OF N-GLYCANS
The biosynthesis of all eukaryotic N-glycans begins on the cytoplasmic face of the ER membrane with the transfer of GlcNAc-P from UDP-GlcNAc to the lipid-like precursor dolichol phosphate (Dol-P) to generate dolichol pyrophosphate N-acetylglucosamine (Dol-P-P-GlcNAc). Fourteen sugars are sequentially added to Dol-P before en bloc transfer of the entire glycan to an Asn-X-Ser/Thr sequon in a protein that is being synthesized and translocated through the ER membrane. The protein-bound N-glycan is subsequently remodeled in the ER and Golgi by a complex series of reactions catalyzed by membrane-bound glycosidases and glycosyltransferases. Many of these enzymes are exquisitely sensitive to the physiological and biochemical state of the cell in which the glycoprotein is expressed. Thus, the populations of sugars attached to each glycosylated asparagine in a mature glycoprotein will depend on the cell type in which the glycoprotein is expressed and on the physiological status of the cell, a status that may be regulated during development and differentiation and altered in disease.
Synthesis of the Dolichol-P-P-Glycan Precursor
Dolichol is a polyisoprenol lipid comprised of five-carbon isoprene units linked linearly in a head-to-tail fashion. Dol-P is used in N-glycan synthesis (Figure 8.2). The number of isoprene units in dolichol varies within cells and between cell types and organisms. For example, the most common yeast dolichol has 14 isoprene units, whereas dolichols from other eukaryotes, including mammals, are longer and may have up to 19 isoprene units. The structure of the N-glycan precursor that is synthesized on Dol-P is shown in Figure 8.3. The enzymes that catalyze each step in the biosynthesis have been identified primarily from studies of mutants of the yeast Saccharomyces cerevisiae. The gene affected by each yeast mutation is known as an ALG gene (for altered in glycosylation) as shown in Figure 8.3.

FIGURE 8.2
Dolichol phosphate (Dol-P). N-Glycan synthesis begins by the transfer of GlcNAc-1-P from UDP-GlcNAc to Dol-P to generate dolichol pyrophosphate N-acetylglucosamine (Dol-P-P-GlcNAc). This reaction is inhibited by tunicamycin.

FIGURE 8.3
Synthesis of dolichol-P-P-GlcNAc2Man9Glc3. Dolichol (red squiggle) phosphate (Dol-P) located on the cytoplasmic face of the ER membrane receives GlcNAc-1-P from UDP-GlcNAc in the cytoplasm to generate Dol-P-P-GlcNAc. Dol-P-P-GlcNAc is extended to Dol-P-P-GlcNAc (more...)
Synthesis of the N-glycan precursor is initiated on the cytoplasmic face of the ER by the transfer of GlcNAc-P from UDP-GlcNAc to membrane-bound Dol-P, forming GlcNAc-P-P-Dol (Figure 8.3). This step is catalyzed by the enzyme GlcNAc-1-phosphotransferase that transfers GlcNAc-1-P from UDP-GlcNAc. Abolition of N-glycosylation in cells and embryos can be achieved by treatment with tunicamycin, an analog of UDP-GlcNAc that inhibits this enzyme. A second N-acetylglucosamine and five mannose residues are subsequently transferred in a stepwise manner from UDP-GlcNAc and GDP-Man, respectively, to generate Man5GlcNAc2-P-P-Dol on the cytoplasmic side of the ER. Each of the sugar additions is catalyzed by a specific glycosyltransferase. Only GlcNAc-1-phosphotransferase transfers a sugar linked to phosphate (N-acetylglucosamine-1-P). All of the other enzymes transfer only the sugar portion of the nucleotide sugar. By a mechanism that is not fully understood, the Man5GlcNAc2-P-P-Dol precursor translocates across the ER membrane bilayer so that the glycan becomes exposed to the lumen of the ER. This translocation is mediated by a “flippase,” which has been genetically linked to the Rtf1 locus in yeast. Man5GlcNAc2-P-P-Dol is extended by the addition of four mannose residues transferred from Dol-P-Man. Assembly of the Dol-P-P-glycan precursor is completed with the addition of three glucose residues donated by Dol-P-Glc. Dol-P-Man and Dol-P-Glc donors are formed on the cytoplasmic side of the ER from GDP-Man and UDP-Glc by transfer of the respective sugar to Dol-P. Dol-P-Man and Dol-P-Glc must also be flipped across the ER bilayer. Mpdu1 protein is necessary for the bioavailability of Dol-P-Man and Dol-P-Glc in the ER for use by the mannosyltransferases and glucosyltransferases that catalyze the synthesis of the mature N-glycan precursor Glc3Man9GlcNAc2-P-P-Dol (Figure 8.3). This 14-sugar glycan is now ready for transfer to asparagine in receptive Asn-X-Ser/Thr sequons of protein regions that have translocated across the ER membrane.
Transfer of the Dolichol-linked Precursor to Nascent Proteins
A multisubunit protein complex in the ER membrane is responsible for catalyzing the transfer of Glc3Man9GlcNAc2 from Dol-P-P to Asn-X-Ser/Thr in newly synthesized regions of proteins as they emerge from the translocon in the ER membrane. The complex that transfers the 14-sugar glycan is termed oligosaccharyltransferase (OST). The OST complex binds to the membrane-anchored Dol-P-P-oligosaccharide and transfers the glycan to nascent protein by cleavage of the high-energy GlcNAc-P bond, releasing Dol-P-P in the process (Figure 8.3). Yeast OST complexes have been purified and are comprised of nine different membrane-bound subunits denoted by their gene names Ost1p, Wbp1p, Swp1p, Ost2p, Ost3p, Ost6p, Ost4p, Ost5p, and Stt3p. Five subunits (Ost1p, Wbp1p, Swp1p, Ost2p, and Stt3p) are essential for yeast viability. Stt3p appears to be the catalytic subunit. Yeast OST exists in three multisubunit complexes: (1) Stt3p-Ost3p-Ost4p, Ost1p-Ost5p; (2) Stt3p-Ost6p-Ost4p, Ost1p-Ost5p; and (3) Wbp1p-Ost2p-Swp1p. All OST subunits are trans-membrane proteins with between one and eight transmembrane domains. Interestingly, a homolog of the STT3 gene has been identified in the prokaryote Campylobacter jejuni and the corresponding protein has been shown to mediate en bloc N-glycosylation of asparagine residues in that organism. Three OST complexes have been identified in mammals. All contain ribophorins I and II, OST48, and DAD1 (defender against apoptotic cell death), which encode proteins related to Ost1p, Swp1p, Wbp1p, and Ost2p, respectively. In addition, mammalian OST contains other associated proteins and one of two Stt3p proteins (A or B), two distinct Stt3p isoforms that are differentially expressed in different cell types. Mammalian OST-I, OST-II, and OST-III differ in their kinetic properties and in their abilities to transfer Dol-P-P-glycans that have fewer than 14 sugars. Such immature N-glycan species are generated in Alg yeast mutants (Figure 8.3) and in patients with congenital disorders of glycosylation (Figure 8.3; see Chapter 42). Thus, N-glycan addition may be differentially regulated at several levels and may vary depending on metabolic conditions.
Early Processing Steps: Glc3Man9GlcNAc2Asn to Man5GlcNAc2Asn
Following the covalent attachment of the 14-sugar oligomannose glycan to Asn-X-Ser/Thr in a polypeptide, a series of processing reactions trims the N-glycan in the ER. The initial steps appear to be conserved among all eukaryotes and are now known to have key roles in regulating glycoprotein folding via interactions with ER chaperones that recognize specific features of the trimmed glycan. Details of the interactions between N-glycans and the chaperones calnexin and calreticulin in glycoprotein folding are presented in Chapter 36. Processing or trimming of Glc3Man9GlcNAc2Asn begins with the sequential removal of glucose residues by α-glucosidases I and II (Figure 8.4). Both glucosidases function in the lumen of the ER, with α-glucosidase I acting specifically on the terminal α1–2Glc and α-glucosidase II sequentially removing the two inner α1–3Glc residues. Removal of glucose residues and the transient re-addition of the innermost glucose during protein folding contribute to the ER retention time of a given glycoprotein. The removal of glucose may be prevented by glucosidase I inhibitors such as castanospermine and deoxynojirimycin (see Chapter 50). In the presence of either of these inhibitors, N-glycans retain the three glucose residues and usually lose one or two mannose residues as they pass through the ER and medial-Golgi, resulting in Glc3Man7–9GlcNAc2 structures on mature glycoproteins. Before exiting the ER, many glycoproteins are acted on by ER α-mannosidase I, which specifically removes the terminal α1–2Man from the central arm of Man9GlcNAc2 to yield a Man8GlcNAc2 isomer (Figure 8.4). A second α-mannosidase I–like protein that lacks enzyme activity is also a resident of the ER. It is called EDEM (ER degradation-enhancing α-mannosidase I–like protein) and it has an important role in the recognition of misfolded glycoproteins, thereby targeting them for ER degradation. The function of EDEM in the quality control of ER glycoproteins is described in Chapter 36. The majority of glycoproteins exiting the ER en route to the Golgi carry N-glycans with either eight or nine mannose residues, depending on whether they have been acted on by ER α-mannosidase I. Some N-glycans in the cis-Golgi retain a glucose residue because of incomplete processing in the ER. A Golgi-resident endo-α-mannosidase cleaves internally between the two mannose residues of the Glcα1–3Manα1–2Manα1–2 moiety of such N-glycans, precisely removing the terminal glucose and the mannnose to which it is attached, thereby generating a different Man8GlcNAc2 isomer to that produced in the ER by α-mannosidase I. In multicellular organisms, trimming of α1–2Man residues continues in the Golgi with the action of α1–2 mannosidases IA, IB, and 1C in the cis-Golgi to give Man5GlcNAc2 (Figure 8.4), a key intermediate in the pathway to hybrid and complex N-glycans (Figure 8.1). However, all N-glycans are not fully processed to Man5GlcNAc2, and those incompletely processed glycans cannot undergo remodeling to form hybrid and complex structures. Some Man5GlcNAc2 may also escape further modification. In these cases, a mature membrane or secreted glycoprotein will carry oligomannose N-glycans of the type Man5–9GlcNAc2. In addition, the action of α-mannosidase I can be blocked by the inhibitor deoxymannojirimycin, resulting in Man8GlcNAc2 on mature glycoproteins. Most mature glycoproteins have some oligomannose N-glycans that were not processed in the cis-Golgi.

FIGURE 8.4
Processing and maturation of an N-glycan. The mature Dol-P-P-glycan, synthesized as described in Figure 8.3, is transferred to Asn-X-Ser/Thr sequons during protein synthesis as proteins are being translocated into the ER. Following transfer of the 14-sugar Glc (more...)
In contrast to multicellular organisms, yeast do not truncate the Man8GlcNAc2 N-glycans that enter the cis-Golgi (see Chapter 21). Instead, they add additional mannose residues to Man8GlcNAc2 to produce oligomannose structures containing many branched mannose residues. Such large yeast mannans are antigenic in humans, and thus yeast is not a good host for the production of recombinant therapeutic glycoproteins unless it is genetically manipulated to generate mammalian-type N-glycans.
Late Processing Steps: From Man5GlcNAc2Asn to Hybrid and Complex N-Glycans
Biosynthesis of hybrid and complex N-glycans (Figure 8.1) is initiated in the medial-Golgi by the action of an N-acetylglucosaminyltransferase called GlcNAcT-I, which adds an N-acetyl-glucosamine residue to C-2 of the mannose α1–3 in the core of Man5GlcNAc2 (Figure 8.4). Once this step has occurred, the majority of N-glycans are trimmed by α-mannosidase II, another resident of the medial-Golgi, which removes the terminal α1-3Man and α1-6Man residues from GlcNAcMan5GlcNAc2 to form GlcNAcMan3GlcNAc2. It is important to note that α-mannosidase II cannot trim the Man5GlcNAc2 intermediate unless it is first acted on by GlcNAcT-I. Once the two mannose residues are removed, a second N-acetylglucosamine is added to C-2 of the mannose α1–6 in the core by the action of GlcNAcT-II to yield the precursor for all biantennary, complex N-glycans. Hybrid N-glycans are formed if the GlcNAcMan5GlcNAc2 glycan is not acted on by α-mannosidase II, leaving the peripheral α1–3Man and α1–6Man residues intact and unmodified in the mature glycoprotein. Incomplete action of α-mannosidase II can result in GlcNAcMan4GlcNAc2 hybrids. Another Golgi mannosidase, discovered in mutant mice lacking functional α-mannosidase II, is termed α-mannosidase IIX and also acts on the GlcNAcMan5GlcNAc2 generated by GlcNAcT-1. Inactivation of both α-mannosidase II and α-mannosidase IIX in the mouse leads to embryos lacking all complex N-glycans. Small oligomannose N-glycans have been found in relatively large amounts in invertebrates and plants. These Man3–4GlcNAc2 N-glycans (called paucimannose N-glycans) are formed from GlcNAcMan3–4GlcNAc2, following the removal of the peripheral N-acetylglucosamine residue by a Golgi hexosaminidase that acts after α-mannosidase II.
The complex N-glycan shown in the medial-Golgi of Figure 8.4 has two antennae or branches initiated by the addition of two terminal N-acetylglucosamine residues. Additional branches can be initiated at C-4 of the core mannose α1–3 (by GlcNAcT-IV) and C-6 of the core mannose α1–6 (by GlcNAcT-V) to yield tri- and tetra-antennary N-glycans (Figure 8.5). Another enzyme, termed GlcNAcT-IX or GlcNAcT-Vb, catalyzes the same reaction as GlcNAcT-V on the C-6 core mannose α1–6, but in contrast to GlcNAcT-V, GlcNAcT-IX/Vb can also transfer N-acetylglucosamine to C-6 of the core mannose α1–3. Another branch can be initiated at C-4 of the core mannose α1–3 by GlcNAcT-VI (see Figure 8.5). Highly branched hepta-antennary structures have so far been found in birds and fish, but not yet in mammals, although genes related to GlcNAcT-VI exist in mammalian genomes.

FIGURE 8.5
Branching of complex N-glycans. The hybrid and mature, biantennary, complex N-glycans shown in Figure 8.4 may contain more branches due to the action of branching N-acetylglucosaminyltranferases in the Golgi. The latter can act only after the prior action (more...)
Complex and hybrid N-glycans may carry a “bisecting” N-acetylglucosamine residue that is attached to the β-mannose of the core by GlcNAcT-III (Figure 8.5). The presence of a bisecting N-acetylglucosamine inhibits trimming by α-mannosidase II and also prevents the actions of GlcNAcT-II, GlcNAcT-IV, and GlcNAcT-V in in vitro assays. However, bi-, tri- and tetra-antennary complex N-glycans with a bisecting N-acetylglucosamine are synthesized when GlcNAcT-III acts after α-mannosidase II and the initiation of branches by GlcNAcT-II, GlcNAcT-IV, and/or GlcNAcT-V. A bisecting N-acetylglucosamine on a biantennary N-glycan is shown in Figure 8.5, and it may be present in all of the more highly branched structures.
Maturation of N-Glycans
Further sugar additions, mostly occuring in the trans-Golgi, convert the limited repertoire of hybrid and branched N-glycans into an extensive array of mature, complex N-glycans. For convenience, this part of the biosynthetic process can be divided into three components: (1) sugar additions to the core, (2) elongation of branching N-acetylglucosamine residues by sugar additions, and (3) “capping” or “decoration” of elongated branches. These processes are summarized below.
- In vertebrate N-glycans, the main core modification is the addition of fucose in an α1–6 linkage to the N-acetylglucosamine adjacent to asparagine in the core (Figure 8.6A). Fucosylation in invertebrate glycoproteins also occurs on this N-acetylglucosamine, but the fucose can be added in α1–3 and/or α1–6 linkages (Figure 8.6B). Invertebrate glycoproteins may have up to four fucose residues on the two N-acetylglucosamines of the N-glycan core (Figure 8.6B). In plants, the fucose is transferred to the asparagine N-acetyl-glucosamine only in α1–3 linkage (Figure 8.6B). The fucosyltransferases involved in transferring fucose to this core N-acetylglucosamine require the prior action of GlcNAcT-I (Figure 8.4). Another common modification of the core, notably in plant and helminth glycoproteins, is the addition of xylose in β1–2 linkage to the β-mannose of the core. This xylosyltransferase also requires the prior action of GlcNAcT-I. Xylose has not been detected in vertebrate N-glycans. A few additional core structures have been identified in mammals, but they appear to be very rare. Two examples are given in Figure 8.6C.
- The majority of complex and hybrid N-glycans (denoted by “R” in Figure 8.7) have elongated branches that are made by the addition of a β-linked galactose residue to the initiating N-acetylglucosamine to produce the ubiquitous building block Galβ1-4GlcNAc, referred to as a type-2 N-acetyllactosamine or“LacNAc”sequence (Figure 8.7A).Antennae can be further lengthened by the sequential addition of N-acetylglucosamine and galactose residues, resulting in tandem repeats of LacNAc (-3Galβ1–4GlcNAcβ1-)n, termed poly-N-acetyllactosamine or polyLacNAc (Figure 8.7A). In a variation, β-linked galactose is added to the C-3 of the N-acetylglucosamine to yield Galβ1–3GlcNAc (Figure 8.7B), referred to as a type-1 N-acetyllactosamine (LacNAc) sequence. In some glycoproteins, β-linked N-acetylgalactosamine is added to N-acetylglucosamine instead of β-linked galactose, yielding antennae with a GalNAcβ1–4GlcNAc (“LacdiNAc”) extension (Figure 8.7C). In contrast to poly-N-acetyllactosamine, which is a relatively common structure, tandem repeats of LacdiNAc and type-1 sequences are uncommon, although poly-N-acetyllactosamine sequences are sometimes terminated with a type-1 unit. The structures and biosynthesis of poly-N-acetyllactosamines are discussed further in Chapter 13.
- The most important “capping” or “decorating” reactions involve the addition of sialic acid, fucose, galactose, N-acetylgalactosamine, and sulfate to the branches described in the preceding paragraph. Capping sugars are most commonly α-linked and therefore protrude away from the β-linked ribbon-like poly-N-acetyllactosamine branches, thus facilitating the presentation of terminal sugars to lectins and antibodies. Many of these structures are shared by N- and O-glycans and by glycolipids, and for this reason their detailed description is presented in Chapter 13.
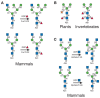
FIGURE 8.6
Modifications of the core of N-glycans. (A) A fucose residue may be transferred to the core of N-glycans after GlcNAcT-I has acted. Thus, hybrid and biantennary, complex N-glycans may have a core fucose. (B) Plants and invertebrates may have additional (more...)

FIGURE 8.7
Elongation of branch N-acetylglucosamine residues of N-glycans. (A) A single N-acetyllactosamine unit is generated when galactose is transferred to a branch N-acetylglucosamine on an N-glycan (R). Further elongation to form poly-N-acetyllactosamine occurs (more...)
The various processes described above potentially yield a myriad of complex N-glycans that differ in branch number, composition, length, capping arrangements, and core modifications. Some examples to illustrate this diversity are shown in Figure 8.8. Many more examples may be found throughout this book.

FIGURE 8.8
Typical complex N-glycan structures found on mature glycoproteins. Symbol Key:

THE SPECIAL CASE OF THE PHOSPHORYLATED N-GLYCANS ON LYSOSOMAL HYDROLASES
Hydrolase enzymes reside in lysosomes where they degrade proteins, lipids, and glycans. Many of these enzymes are targeted to lysosomes by a specialized trafficking pathway that requires the generation of phosphorylated N-glycans. The phosphorylation step occurs in the cis-Golgi and involves the transfer of GlcNAc-1-P to C-6 of mannose residues of oligomannose N-glycans on lysosomal hydrolases (Figure 8.4). A glycosidase in the trans-Golgi removes the N-acetylglucosamine to generate Man-6-P residues. Such residues are recognized by lectin receptors (termed Man-6-P receptors) that transport the lysosomal hydrolase to an acidified compartment where it is released from the receptor and ultimately ends up in a lysosome. The details of the synthesis of the Man-6-P recognition marker on lysosomal hydrolases and the trafficking pathway are presented in Chapter 30.
TRANSFERASES AND TRANSPORTERS IN N-GLYCAN SYNTHESIS
The glycosyltransferases that operate in the ER are mainly multitransmembrane proteins that are woven into the ER membrane. In contrast, the glycosyltransferases in Golgi compartments are generally type II membrane proteins with a small cytoplasmic amino-terminal domain, a single transmembrane domain, and a large lumenal domain that has an elongated stem region and a globular catalytic domain (see Chapter 5). The stem region can be cleaved, releasing the catalytic domain into the lumen of the Golgi and allowing its secretion. Thus, extracellular soluble forms of many glycosyltransferases exist in tissues and sera. At least one soluble form has a unique function. Soluble GlcNAcT-V is an angiogenic factor in tumors. However, extracellular soluble glycosyltransferases are not expected to function as transferases because nucleotide sugars are not known to be present extracellularly. Nucleotide sugars are synthesized in the cytoplasm, except for CMP-sialic acid, which is synthesized in the nucleus (see Chapter 4). They are subsequently concentrated in the appropriate compartment following transport across the membrane by specialized nucleotide sugar transporters. Specific genes encode the nucleotide sugar transporters, which translocate CMP-sialic acid, UDP-Gal, UDP-GlcNAc, GDP-Fuc, and other nucleotide sugars. A few of these transporters can transport more than one nucleotide sugar. Each transporter is a multitransmembrane protein and usually contains ten membrane-spanning domains.
N-LINKED GLYCOPROTEINS COMPRISE MANY GLYCOFORMS
As mentioned above, glycoproteins are heterogeneous with respect to their content of N-glycans and often have a range of different N-glycans on a particular Asn-X-Ser/Thr N-glycosylation sequon. For example, 58 different complex N-glycan structures have been identified at one N-glycan site in mouse zona pellucida glycoprotein 3. Furthermore, if there is more than one Asn-X-Ser/Thr sequon per molecule, different molecules in a population may have different subsets of N-glycans on different sequons. This is referred to as site-specific glycan heterogeneity or microheterogeneity. A homogeneous glycoprotein component of a population is called a glycoform. It appears that the protein sequence or conformation can cause N-glycan diversity, presumably by affecting substrate availability for Golgi glycosidases or glycosyltransferases. Other factors affecting N-glycan heterogeneity include nucleotide sugar metabolism, transport rates of the glycoprotein through the lumen of the ER and Golgi, and the proximity of an N-glycan attachment sequon to a transmembrane domain. Also, localization of glycosyltransferases within subcompartments of the Golgi can determine which enzymes encounter N-glycan substrates first. With respect to the last point, it is important to note that processing enzymes often compete for the same substrate and that most glycosyltransferases and glycosidases require the prior actions of other glycosyltransferases and glycosidases before they can carry out their reactions.
FUNCTIONS OF N-GLYCANS
Determining the functions of N-glycans may be accomplished by the use of inhibitors of N-linked glycosylation such as tunicamycin; inhibitors of N-glycan processing such as castanospermine, deoxynojirimycin, and swainsonine; or the generation of mutants in a gene that codes for a glycosylation activity in model organisms such as yeast, cultured mammalian cells, Drosophila, C. elegans, zebrafish, and mouse. The various chemical inhibitors of N-glycan synthesis are discussed in Chapter 50. Many yeast mutants in the synthesis and initial processing of N-glycans are identified in Figure 8.3, and three of the many mutants of cultured cells with altered glycosylation are identified in Figure 8.4 and described in detail in Chapter 46. Mutant cells or organisms with an altered N-glycosylation ability shed enormous insights not only into the biological functions of N-glycans, but also into their contributions to the biochemical properties of a glycoprotein in terms of structure, susceptibility to proteases, and antigenicity. In addition, mutant cells and organisms allow the glycosylation pathways that operate in vivo to be defined. A cell or organism with a loss-of-function mutation often accumulates the biosynthetic intermediate that is the substrate of the activity lost by the mutant. Gain-of-function mutations reveal alternative pathways or glycosylation reactions that may occur. Thus, the study of cells and organisms mutated in a specific gene that affects N-glycosylation has been a major source of information regarding the functions of N-glycans. N-glycan functions have also been determined from the features of human diseases that arise from a defect in N-glycan synthesis. An important group of such diseases is called congenital disorders of glycosylation (CDG), and these diseases are described in Chapter 42.
Mouse mutants in particular have provided enormous insights into the functions of individual sugars present in N-glycans as well as the functions of whole classes of N-glycans. Thus, deletion of the Mgat1 gene that encodes GlcNAcT-I prevents the synthesis of complex and hybrid N-glycans, and Man5GlcNAc2 is found at all complex and hybrid N-glycan sites. Whereas the absence of GlcNAcT-I does not affect the viability or growth of Lec1 cultured cells (Figure 8.4), elimination of GlcNAcT-I in the mouse results in death during embryonic development (see Chapter 25). The complex N-glycans that fail to be synthesized in mice lacking GlcNAcT-I, GlcNAcT-V, GlcNAcT-IVb, and FucT-VIII are important in retaining growth factor and cytokine receptors at the cell surface, probably through interactions with glycan-binding proteins such as galectins or cytokines such as transforming growth factor β. Cell-surface receptors and a glucose transporter lacking branches of a complex N-glycan have a shorter residence time on the cell surface and their signaling is attenuated. Deletion of genes encoding sialyltransferases, fucosyltransferases, or branching N-acetylglucosaminyltranferases other than GlcNAcT-I has generally produced viable mice with defects in immunity or neuronal cell migration, emphysema of the lung, or inflammation. N-glycans may carry the sugar determinants recognized by selectins that mediate cell–cell interactions important for leukocyte extravasation from the blood stream and regulate lymphocyte homing to lymph nodes (see Chapter 31). N-Glycans are known to become more branched when cells become cancerous, and this change facilitates cancer progression (see Chapter 44). Tumors formed in mice lacking GlcNAcT-V or GlcNAcT-III (Figure 8.5) may be retarded in their progression. Thus, certain glycosyltransferases may be appropriate targets for the design of cancer therapeutics.
FURTHER READING
- Waechter CJ, Lennarz WJ. The role of polyprenol-linked sugars in glycoprotein synthesis. Annu Rev Biochem. 1976;45:95–112. [PubMed: 786163]
- Snider MD, Sultzman LA, Robbins PW. Transmembrane location of oligosaccharide-lipid synthesis in microsomal vesicles. Cell. 1980;21:385–392. [PubMed: 6250720]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. [PubMed: 3896128]
- Herscovics A. Importance of glycosidases in mammalian glycoprotein biosynthesis. Biochim. Biophys. Acta. 1999a;1473:96–107. [PubMed: 10580131]
- Herscovics A. Processing glycosidases of Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1999b;1426:275–285. [PubMed: 9878780]
- Berninsone PM, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 2000;10:542–547. [PubMed: 11042451]
- Schachter H. The joys of HexNAc. The synthesis and function of N- and O-glycan branches. Glycoconj J. 2000;17:465–483. [PubMed: 11421343]
- Schenk B, Fernandez F, Waechter CJ. The ins(ide) and outs(ide) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 2001;11:61R–70R. [PubMed: 11425794]
- Lowe JB, Marth JD. A genetic approach to mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. [PubMed: 12676797]
- Freeze HH, Aebi M. Altered glycan structures: The molecular basis of congenital disorders of glycosylation. Curr Opin Struct Biol. 2005;15:490–498. [PubMed: 16154350]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. [PubMed: 16317064]
- Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. [PubMed: 17418791]
- DISCOVERY AND BACKGROUND
- MAJOR STRUCTURAL CLASSES AND NOMENCLATURE
- PREDICTING SITES OF N-GLYCOSYLATION
- ISOLATION, PURIFICATION, AND ANALYSIS
- SYNTHESIS OF N-GLYCANS
- THE SPECIAL CASE OF THE PHOSPHORYLATED N-GLYCANS ON LYSOSOMAL HYDROLASES
- TRANSFERASES AND TRANSPORTERS IN N-GLYCAN SYNTHESIS
- N-LINKED GLYCOPROTEINS COMPRISE MANY GLYCOFORMS
- FUNCTIONS OF N-GLYCANS
- FURTHER READING
- Review N-Glycans.[Essentials of Glycobiology. 2015]Review N-Glycans.Stanley P, Taniguchi N, Aebi M. Essentials of Glycobiology. 2015
- Review N-Glycans.[Essentials of Glycobiology. 2022]Review N-Glycans.Stanley P, Moremen KW, Lewis NE, Taniguchi N, Aebi M. Essentials of Glycobiology. 2022
- Review O-GalNAc Glycans.[Essentials of Glycobiology. 2009]Review O-GalNAc Glycans.Brockhausen I, Schachter H, Stanley P. Essentials of Glycobiology. 2009
- Review Lysosomal metabolism of glycoproteins.[Glycobiology. 2005]Review Lysosomal metabolism of glycoproteins.Winchester B. Glycobiology. 2005 Jun; 15(6):1R-15R. Epub 2005 Jan 12.
- N-glycosylation at one rabies virus glycoprotein sequon influences N-glycan processing at a distant sequon on the same molecule.[Glycobiology. 2005]N-glycosylation at one rabies virus glycoprotein sequon influences N-glycan processing at a distant sequon on the same molecule.Wojczyk BS, Takahashi N, Levy MT, Andrews DW, Abrams WR, Wunner WH, Spitalnik SL. Glycobiology. 2005 Jun; 15(6):655-66. Epub 2005 Jan 26.
- N-Glycans - Essentials of GlycobiologyN-Glycans - Essentials of Glycobiology
- Mixture/Component Compounds for PubChem Compound (Select 657298) (0)PubChem Compound
- Taxonomy for PubChem Compound (Select 6870646) (9)Taxonomy
- ribulose-1,5-bisphosphate carboxylase/oxygenase form I, partial [delta proteobac...ribulose-1,5-bisphosphate carboxylase/oxygenase form I, partial [delta proteobacterium SCGC AAA240-M14]gi|322511688|gb|ADX05613.1|Protein
Your browsing activity is empty.
Activity recording is turned off.
See more...

