NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009.

Essentials of Glycobiology. 2nd edition.
Show detailsThis chapter presents an overview of the dynamic modification of serine or threonine hydroxyl moieties on nuclear and cytoplasmic proteins by β-linked N-acetylglucosamine, termed O-β-GlcNAc or simply O-GlcNAc.
HISTORICAL BACKGROUND
O-GlcNAc is distinct from all other common forms of protein glycosylation in several major respects (Figure 18.1). (1) It occurs exclusively within the nuclear and cytoplasmic compartments of the cell. (2) The GlcNAc is generally not elongated or modified to form more complex structures. (3) It is attached and removed multiple times in the life of a polypeptide, often cycling rapidly and at different rates at different sites on a polypeptide. In most ways, O-GlcNAcylation is more similar to protein phosphorylation than it is to “classical” protein glycosylation. Research on O-GlcNAc is particularly relevant to chronic human diseases including diabetes, cardiovascular disease, neurodegenerative disorders, and cancer. A long-held dogma, still promulgated in many cell biology and biochemistry textbooks today, is that protein glycosylation does not occur within the cytoplasmic and nuclear compartments of the cell. This view is apparently true for typical N-glycans and O-glycans that are attached to protein by glycosyltransferases with their active sites within the lumenal compartments of the ER and Golgi (see Chapters 3 and 8–12). However, it is now clear that certain specialized types of protein glycosylation do, indeed, occur within the nucleus and the cytoplasm (see Chapter 17). In fact, during the past two decades, it has become clear that O-GlcNAc is one of the most abundant posttranslational modifications within the nucleocytoplasmic compartments of all metazoans including plants, animals, and the viruses that infect them (Figures 18.1 and 18.2 and Table 18.1).

FIGURE 18.1
(A) O-Linked β-N-acetylglucosamine (O-GlcNAc) is a dynamic protein modification abundant within the nucleus and cytoplasm. The O-GlcNAc transferase (OGT) attaches O-GlcNAc to proteins at specific serine or threonine residues. O-GlcNAcase hydrolyzes (more...)

FIGURE 18.2
(A) O-GlcNAcylated proteins occur in many different cellular compartments. Found mostly within the nucleus, they are also in the cytoplasm and within viruses that infect eukaryotic cells. The subcellular distribution of O-GlcNAc is similar to that of (more...)
TABLE 18.1
Some O-GlcNAcylated proteins
O-GlcNAc was discovered in 1983, when purified bovine milk galactosyltransferase and its radiolabeled donor substrate (UDP-[3H]galactose) were used to probe for GlcNAc-terminating glycoconjugates in living murine thymocytes, splenic B- and T-lymphocytes, and macrophages. Galactosyltransferase is a Golgi glycosyltransferase that attaches galactose in a β1-4 linkage to almost any terminal N-acetylglucosamine residue. Contrary to expectations, product analyses, including release from protein by alkali-induced β-elimination and resistance to cleavage by peptide-N-glycosidase F (PNGase F; see Chapters 8 and 47), showed that most of the galactosylated glycans existed within the cell as single O-linked GlcNAc moieties. Studies of the subcellular localization of O-GlcNAc in rat liver established that it is most abundant within chromatin, most concentrated on the nuclear pores of the nuclear envelope, and also present within the cytoplasm of the cell. There is so far no evidence for O-GlcNAc on the regions of polypeptides that reside on the outside of cells or within lumenal compartments. Galactosyltransferase in combination with donor substrates of either radiolabeled UDP-galactose or chemically modified UDP-galactose, to which fluorescent or biotin tags can be attached, still represents one of the best methods for detecting and studying O-GlcNAc. More recent studies have used metabolic incorporation of N-azidoacetylglucosamine in living cells, combined with chemical tagging, affinity isolation, and mass spectrometry, to identify larger numbers of O-GlcNAc–modified proteins. Unlike many other methods, these covalent probes for O-GlcNAc permit direct chemical and enzymatic analyses of the products formed.
O-GlcNAc IS A HIGHLY DYNAMIC MODIFICATION
Unlike the relatively stable nature of mature N- and O-glycans on glycoproteins, O-GlcNAc cycles rapidly on and off most proteins because of the combined action of O-GlcNAc transferase (sometimes abbreviated to OGT) and O-GlcNAcase (Figure 18.3). Early studies showed that mitogen or antigen activation of lymphocytes rapidly decreased O-GlcNAcylation of many cytoplasmic proteins, but concomitantly increased O-GlcNAcylation of many nuclear proteins. Likewise, neutrophils were shown to rapidly modulate O-GlcNAcylation of several proteins in response to chemotactic agents. Pulse-chase analyses have revealed that O-GlcNAc residues on small heat-shock protein in lens (α-crystallin) and on intermediate filament proteins (cytokeratins) turn over more rapidly than the polypeptide chains to which they are attached. Similar studies on several proteins have confirmed that O-GlcNAc cycles, sometimes very rapidly, depending on the protein as well as the site on the protein to which it is attached. Recent proteomic studies that have quantified O-GlcNAc cycling in response to external signals (e.g., insulin, high glucose, or cellular stress) and/or inhibitors of O-GlcNAcase have established that the rates of O-GlcNAc cycling are similar to those often seen for protein phosphorylation, with half-lives of 1 minute or even less. It is also now clear that O-GlcNAc levels change during the cell cycle and that O-GlcNAc is involved in cell-cycle progression. However, on some proteins (e.g., nuclear pore proteins) O-GlcNAc cycles slowly or only in response to external signals that remain to be identified.

FIGURE 18.3
O-GlcNAc cycling is regulated by the concerted action of O-GlcNAc transferase (OGT) and O-GlcNAcase. OGT has two distinct domains: the transferase domain, which contains the catalytic site and is related to glycogen phosphorylase, and the tetratricopeptide (more...)
Why Did O-GlcNAc Remain Undetected for So Long?
As shown in Figures 18.1 and 18.2, O-GlcNAc is found on hundreds or even thousands of nucleocytoplasmic proteins. In fact, many of the most heavily studied proteins in biology are O-GlcNAcylated (Table 18.1). Yet most researchers devoted to studying the structure and functions of these same proteins remain surprisingly unaware of the presence of O-GlcNAc. So why did O-GlcNAc remain undetected until 1983, and why is it still largely unseen and unconsidered by investigators studying transcription, signaling, and the cytoskeleton? First, unlike charged modifcations (e.g., phosphate), addition and removal of O-GlcNAc generally does not affect the migration of polypeptides on SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) or even on high-resolution 2D gels. A small shift in protein migration might be seen if the O-GlcNAc residues are highly clustered or if the protein is extensively O-GlcNAcylated at multiple sites (e.g., p62 nuclear pore protein or Sp1 transcription factor). Second, all cells contain high levels of hydrolases, including abundant lysosomal hexosaminidases and nucleocytoplasmic β-N-acetylglucosaminidases, that rapidly remove O-GlcNAc from intracellular proteins when the cell is damaged or lysed. Thus, O-GlcNAc is often lost during the isolation of a protein. Third, O-GlcNAc is particularly difficult to detect by physical techniques such as mass spectrometry, because it often occurs at substoichiometric amounts at different sites on a protein and readily falls off the polypeptide during the ionization process in a mass spectrometer. In both electrospray mass spectrometry and in matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry analyses of a mixture of unmodified and O-GlcNAc–modified peptides, not only is the O-GlcNAc lost during ionization, but the signal from the O-GlcNAc–modified peptides that remain is nearly, or even entirely, suppressed by the presence of the unmodified peptide. However, the recent development of monoclonal antibodies that detect O-GlcNAc and the invention of more sophisticated mass spectrometric methods, when combined with new potent inhibitors of β-N-acetylglucosaminidases, have substantially improved our ability to detect O-GlcNAc on proteins.
O-GlcNAc IS UBIQUITOUS AND ESSENTIAL IN METAZOANS
To date, nucleocytoplasmic O-β-GlcNAc has been found in all multicellular organisms investigated, ranging from filamentous fungi, worms, insects, and plants to humans. However, O-GlcNAc and the enzymes that control its cycling (see below) do not appear to exist in yeast, such as Saccharomyces cerevisiae or Schizosaccharomyces pombe. O-α-GlcNAc appears to be a common modification of cell-surface and extracellular proteins in protozoa, where it is attached by enzymes structurally related to the lumenal protein:O-GalNAc transferases (ppGalNAc-Ts) in mammals (see Chapter 9). However, it remains unclear whether nucleocytoplasmic O-β-GlcNAc occurs within either free-living or parasitic protozoa. For example, the known genes that encode the O-GlcNAc transferase and O-GlcNAcase are apparently absent from the genome of the parasitic flagellated protozoan Trypanosoma brucei (see Chapter 40). It now appears that earlier studies reporting O-GlcNAc in protozoa were, in fact, based mostly on the presence of O-α-GlcNAc on extracellular proteins, which appears to serve the same functions as mucin-type α-GalNAc O-glycans in higher organisms, perhaps because many protozoa lack the C-4 epimerase required to convert UDP-GlcNAc to UDP-GalNAc (see Chapter 4). Genetic and subsequent biochemical studies in Arabidopsis showed that the genes SPY and SECRET AGENT regulate growth hormone (gibberellic acid) signaling, and later studies established that both of these genes encode O-β-GlcNAc transferases. Mutations in either SPY or SECRET AGENT cause severe growth phenotypes, but they are not lethal. However, simultaneous mutation of both genes is lethal. Studies in rice, potatoes, and other plants have also indicated that O-GlcNAcylation is important for growth regulation.
Unlike plants, mammals and insects appear to have only a single gene encoding the catalytic subunit of the O-GlcNAc transferase (OGT). Using Cre-LoxP conditional gene disruption in mice, OGT was shown to be required for the viability of embryonic stem cells. Tissue-targeted disruption in mice and disruption of OGT expression in cell culture have established that O-GlcNAcylation is essential for viability at the single-cell level in mammalian cells. Disruption of OGT in the worm Caenorhabditis elegans causes defective carbohydrate metabolism and abnormalities in dauer formation, a metabolically inactive larval form of the worm that is long lived and stress resistant, analogous to spores in bacteria. O-GlcNAc–modified proteins have also been found on many viruses that infect metazoans (e.g., adenovirus, SV40, cytomegalovirus, rotovirus, baculovirus, plum pox, HIV, and others). In viruses, instead of being localized on the outside capsid of the virus where “classical” N- or O-glycans are found (see Chapter 39), O-GlcNAc is found deep within the viruses, on tegument and other regulatory proteins, close to the nucleic acid components. Thus far, at least one pathogenic bacterium (Clostridium novyi) has been found to exploit the O-GlcNAc modification. The α toxin of C. novyi is an O-GlcNAc transferase (no homology to the mammalian enzyme) that kills cells by attaching an O-GlcNAc residue to GTP-binding sites on the small G proteins such as Rho, Rac, and Ran that regulate the cytoskeleton. Some of these G proteins are normally O-GlcNAcylated by an endogenous O-GlcNAc transferase, but the sites of attachment of O-GlcNAc are not known.
O-GlcNAc Is Found on a Wide Range of Proteins
Studies from several laboratories have described O-GlcNAcylated proteins from virtually all cellular compartments representing nearly all functional classes of protein (Figure 18.2). In short, O-GlcNAc occurs everywhere that Ser(Thr)-O-phosphorylation has been found. O-GlcNAc is particularly abundant within the nucleus, where it occurs on the transcriptional regulatory machinery including RNA polymerase II catalytic subunit carboxy-terminal domain (CTD) and myriad basal and specialized RNA polymerase II transcription factors. About one quarter of O-GlcNAcylated proteins are transcription or translation regulatory factors; nuclear pore proteins are among the most heavily O-GlcNAcylated proteins. O-GlcNAcylation also appears to be particularly abundant on proteins involved in signaling, stress responses, and energy metabolism. It also occurs on many cytoskeletal regulatory proteins, such as those regulating actin assembly (e.g., vinculin, talin, vimentin, and ankyrin) and tubulin assembly (e.g., microtubule-associated proteins [MAPs], dynein, and tau). Even α-tubulin itself is dynamically modified by O-GlcNAc, but stoichiometry appears to be low. Intermediate filaments, such as cytokeratins and neurofilaments in brain, are also heavily modified by O-GlcNAc. To date, more than 600 O-GlcNAcylated proteins have been identified, but this large number probably still represents only a portion of the most abundant proteins that are dynamically modified within the cell. Table 18.1 shows a partial list of some of the known O-GlcNAcylated proteins.
Individual sites modified by O-GlcNAc have been identified on several proteins (Table 18.2). About half of the mapped sites have a proline and valine residue on either side of the modified hydroxy amino acid. The “PVS” motif is similar to that recognized by proline-directed kinases, such as glycogen synthase kinase-3β (GSK3β). The other O-GlcNAc sites seemingly have little in common at the primary sequence level, but they have sequences similar to those also recognized by different kinases. Many of the identified O-GlcNAc sites have high “PEST” scores, a sequence motif that is associated with rapid degradation of a protein, and a few studies suggest that O-GlcNAcylation within the PEST sequence prevents rapid degradation of the protein.
TABLE 18.2
Some sites of O-GlcNAc attachment
O-GlcNAc HAS A COMPLEX DYNAMIC INTERPLAY WITH O-PHOSPHATE
O-GlcNAc and O-phosphate exhibit a complex interplay on signaling, transcriptional, and cytoskeletal regulatory proteins within the cell (Figure 18.4). In fact, a major enzyme that removes O-phosphate, protein phosphatase 1 catalytic subunit (PP1c), is in a dynamic complex with OGT, indicating that, in many cases, the same enzyme complex both removes O-phosphate and concomitantly attaches O-GlcNAc (Figure 18.5). The microtubule regulatory protein tau is abundantly phosphorylated in the early stages of brain development, but throughout adult life it is apparently extensively O-GlcNAcylated and not significantly phosphorylated. However, in the brains of patients with Alzheimer’s disease, tau becomes less O-GlcNAcylated and more extensively phosphorylated, leading to the formation of intraneuronal “tangles” composed of paired helical filamentous (PHF)-tau, which are characteristic of this disease. The CTD repeat domain of RNA polymerase II is alternately extensively O-GlcNAcylated (with as many as three O-GlcNAc residues per repeat) and phosphorylated. At some time during the transcription cycle, O-GlcNAc on the CTD is removed and replaced with O-phosphate, initiating the elongation phase of transcription. In vitro studies have shown that synthetic peptides with up to ten CTD repeats (comprising 70 amino acids) cannot be phosphorylated by CTD kinases if they contain even a single O-GlcNAc moiety. Likewise, these CTD peptides cannot be O-GlcNAcylated if even one of the repeats contains a single O-phosphate residue, despite the many potential modification sites available on the same or other repeat sequences.
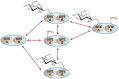
FIGURE 18.4
O-GlcNAcylation exhibits a complex dynamic interplay with O-phosphorylation. The example shown depicts the complexity created by two potential sites on a protein. On some proteins, O-GlcNAc and O-phosphate compete for the same site. On others, their respective (more...)

FIGURE 18.5
Protein phosphatase 1 catalytic subunit (PP1c), a key enzyme that removes O-phosphate residues from many important regulatory proteins, exists in a complex with O-GlcNAc transferase, which attaches O-GlcNAc. This suggests that the same enzyme complex (more...)
O-GlcNAc occurs reciprocally on several transcription factors important to cancer. For example, Thr-58, located within the transactivation domain of the c-myc proto-oncogene, which is the major site mutated in human lymphomas, is alternatively phosphorylated by GSK3β kinase and O-GlcNAcylated by OGT. Phosphorylation at Thr-58 is the culmination of a hierarchical signaling cascade penultimately involving a “priming” phosphorylation reaction characteristic of many GSK3β substrates. First, Ser-62 on c-Myc is phosphorylated by the growth factor kinase ERK, which creates a recognition site that allows Thr-58 of c-Myc to be phosphorylated by GSK3β. Studies with site- and modification-specific antibodies have shown that in nongrowing cells, Thr-58 of c-Myc is modified by O-GlcNAc. However, after applying a growth stimulus, O-GlcNAc is very rapidly removed and replaced by O-phosphate at this site.
In a similar fashion, O-GlcNAcylation of Ser-16 on estrogen receptor β causes it to become less transcriptionally active, but more long lived in the cell. In contrast, O-phosphorylation of Ser-16 causes estrogen receptor β to become more transcriptionally active, but degraded much more rapidly.
Hyper-O-GlcNAcylation, as occurs in diabetes, also blocks phosphate-dependent activation of some proteins. For example, the critical AKT kinase regulatory site on endothelial nitric oxide synthase (eNOS) is occupied by an O-GlcNAc moiety in individuals with diabetes, preventing activation of the enzyme. This O-GlcNAc–mediated block in eNOS activation is associated with the vascular complications and erectile dysfunction seen in patients with diabetes.
On some proteins (e.g., casein kinase II), O-GlcNAc and O-phosphate occur at separate but adjacent sites, yet they still appear to be reciprocal with each other. However, on other proteins, the relationship between O-GlcNAc and O-phosphate dynamics remains unclear. For example, on cytokeratins, O-GlcNAcylation and O-phosphorylation appear to be independently regulated, but may occur mutually exclusively on completely different subsets of the same polypeptides.
Thus, it is now clear that the dynamic interplay between O-GlcNAcylation and O-phosphorylation is widespread and quite complex (Figure 18.4), requiring us to modify current dogma with respect to cellular signaling. We can no longer view the dynamic regulation of signaling, transcription, and other cellular processes simply as binary “on” or “off” systems controlled by the addition and removal of O-phosphate, but rather we must view these processes as an “analog” system in which the dynamic interplay between these two (and perhaps other) modifications, often at multiple sites on a protein, creates an enormous range of highly dynamic structural and functional molecular possibilities.
ENZYMES CONTROLLING O-GlcNAc CYCLING
Even though O-GlcNAc cycles like O-phosphate and it often even competes for the same site, O-GlcNAc regulation differs remarkably from the regulation of protein phosphorylation by specific kinases. Mammalian genomes encode hundreds of site-specific kinase catalytic subunits that are each independently regulated to act like “microswitches” controlling the signaling circuits of the cell. In contrast, mammalian genomes, thus far, have been found to encode only a single, very highly conserved, catalytic subunit for each of the enzymes OGT and O-GlcNAcase (Figure 18.3). Current dogma dictates that different stimuli cause specific phosphate cycling to change on only a few proteins within a signaling cascade or functional complex. However, in the case of O-GlcNAc, different stimuli cause rapid changes in O-GlcNAc cycling concomitantly on many proteins, presumably even in different pathways and complexes. Thus, the dynamics of O-GlcNAcylation appear to act at a “global” level, whereas the dynamics of O-phosphorylation appear to act at a local level. However, although both OGT and O-GlcNAcase each have only a single catalytic subunit, they actually exist within the cell as a multitude of different holoenzymes in which their catalytic subunits are noncovalently bound to a myriad of accessory proteins that appear to control their targeting specificities (Figure 18.6).
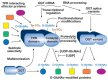
FIGURE 18.6
O-GlcNAc transferase (OGT) is regulated by multiple complex mechanisms, including transcriptional regulation of its expression, differential mRNA splicing, proteolytic processing, posttranslational modification, and multimerization with itself and other (more...)
O-GlcNAc Transferase
O-GlcNAc transferase (OGT; uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase) catalyzes the addition of N-acetylglucosamine from UDP-GlcNAc to specific serine or threonine residues to form a β-glycosidic linkage. The enzyme was first identified and purified from rat liver and subsequently cloned from rat, human, C. elegans, and other organisms. OGT resides on the X chromosome (Xq13 in humans) near the centromere and is among the most highly conserved proteins from worms to man. As described above, OGT is required for single-cell viability in mammals and plants, but not in C. elegans. In rat liver, OGT appears to be a heterotrimer of two 110-kD subunits and one 78-kD subunit. The 78-kD subunit is most abundant in kidney, muscle, and liver, but in most tissues such as the brain and pancreas the enzyme contains only the 110-kD subunits. The functional significance of these variants is unclear. Also, a unique mitochondrial form of OGT has been described. OGT is present in all cells, but is most abundant by far in the glucose-sensing cells (α and β cells) of the pancreas and also in the brain. OGT has two distinct domains separated by a putative nuclear localization sequence. The amino terminus of each OGT subunit contains up to 11.5 tetratricopeptide repeats (TPRs) that serve as protein:protein interaction domains and represent the sites to which most of the OGT targeting/regulatory proteins bind. The crystal structure of human TPRs of OGT show that the repeats occur as stacked α-helical domains, forming a “tube-like” structure with a remarkable structural similarity to the TPR region of the nuclear transport protein importin-α, suggesting that targeting of OGT occurs in a manner similar to importin-α’s mechanism of transporting proteins across the nuclear envelope. The TPRs of OGT are also required for the multimerization of OGT subunits. The carboxy-terminal domain of OGT appears to have evolved from the glycogen phosphorylase superfamily of enzymes, and it contains the UDP-GlcNAc-binding and catalytic sites of the enzyme.
The regulation of OGT is quite complex and still not well understood (Figure 18.6). OGT is itself O-GlcNAcylated and also tyrosine phosphorylated. Tyrosine phosphorylation appears to activate the enzyme, but the role of O-GlcNAc on OGT is not yet clear. Purified or recombinant OGT modifies small synthetic peptides based on known sites from O-GlcNAcylated proteins (Table 18.2), but OGT appears to require accessory proteins bound to the TPRs to modify full-length protein substrates efficiently. Yeast two-hybrid analyses have identified many of these OGT targeting proteins. The best characterized is OIP106, which is involved in targeting OGT to RNA polymerase II–containing transcriptional machinery. However, the most important factor regulating overall OGT activity within the cell is the level of its donor substrate UDP-GlcNAc. The concentrations of cellular UDP-GlcNAc are exquisitely sensitive to carbohydrate, fatty acid, energy, and nitrogen metabolic fluxes (Figure 18.7), making it an ideal metabolic sensor. In cells, 2–5% of all glucose use occurs through the hexosamine biosynthetic pathway, which culminates in the biosynthesis of UDP-GlcNAc (see Chapter 4). Upon transfer of GlcNAc to proteins, UDP is released, which is a potent feedback inhibitor of OGT. Under conditions during which UDP is rapidly removed (as occurs within the cell), OGT activity is dependent on the UDP-GlcNAc level over a remarkable range of concentrations (from the low nM range to well over 50 mM). The concentration of UDP-GlcNAc in a cell typically ranges from approximately 0.1 to 1 mM, second only to ATP for high-energy small molecules. Thus, rapid changes in UDP-GlcNAc concentrations may serve as a nutrient sensor by directly affecting the extent of O-GlcNAcylation of regulatory proteins.
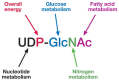
FIGURE 18.7
UDP-GlcNAc is the donor for O-GlcNAc transferase (OGT) and an ideal sensor of the metabolic status of the cell. In cells, 2–5% of glucose is metabolized to form UDP-GlcNAc. Unlike other sugar nucleotides, UDP-GlcNAc levels (which are second only (more...)
O-GlcNAcase
Nucleocytoplasmic β-N-acetylglucosaminidase (O-GlcNAcase) was first identified as a neutral cytosolic hexosaminidase (referred to as “hexosaminidase C”) and often compared to the better-studied lysosomal hexosaminidases A and B. O-GlcNAcase was purified from rat kidney and bovine brain, and the human gene was cloned using peptide sequence information. The O-GlcNAcase gene was found to be identical to MGEA5, a putative hyaluronidase genetically identified because of its association with meningiomas. The O-GlcNAcase gene maps to the late-onset Alzheimer’s disease locus in humans (10q24.1) and lies adjacent to the gene for insulin-degrading enzyme. Sequence analyses suggest that O-GlcNAcase is a bifunctional enzyme (Figure 18.3). The amino terminus of O-GlcNAcase encodes the glycosidase domain, whereas the carboxyl terminus shows homology to the GCN5 HAT family, which specifically acetylates histones to activate gene expression. O-GlcNAcase has both HAT and O-GlcNAcase activities in vitro. During the latter stages of apoptosis (regulated cell death), the executioner caspase (caspase-3) cleaves O-GlcNAcase to separate the HAT and O-GlcNAcase domains. Like OGT, O-GlcNAcase also interacts in a dynamic fashion with a very large number of cellular proteins, but the location of interaction sites on the enzyme have not been mapped. O-GlcNAcase and OGT are often within the same functional complex, particularly in transcription complexes, where both enzymes are found with transcriptional regulators, such as the transcriptional repressor mSin3A. Thus, both enzymes appear to be tightly regulated to prevent futile cycling of O-GlcNAc.
BIOLOGICAL FUNCTIONS OF O-GlcNAc
Like O-phosphorylation, the specific functions of O-GlcNAc depend on the protein and sites to which the glycan is attached. However, on the basis of many studies, some generalities are emerging. As discussed above, one of the major functions of O-GlcNAc is to prevent O-phosphorylation and, by doing so, to modulate signaling and transcription in response to cellular nutrients or stress.
O-GlcNAc Regulates Transcription and Translation
O-GlcNAc directly regulates the activities of a variety of transcription factors including Sp1, estrogen receptors, STAT5, NF-κB, p53, YY1, Elf-1, c-Myc, Rb, PDX-1, CREB, fork-head, and others. As described above, RNA polymerase II and the basal transcription factors are also extensively modified by O-GlcNAc. O-GlcNAc modification has been reported to suppress or enhance transcription, depending on the promoter and other associated proteins. Some examples are outlined below.
- Hyperglycosylation of Sp1 occurs in diabetic states and this modification enhances the transcription of some genes characteristic of this disease, while suppressing the transcription of others. For instance, Sp1 with high O-GlcNAc drives the transcription of plasminogen activator and extracellular matrix proteins thought to have an important role in diabetes-associated cardiovascular disease.
- Increased O-GlcNAc contributes to impaired calcium cycling in the heart and to diabetic cardiomyopathy by reducing the transcription of a sarcoplasmic reticulum (SR) calcium ATPase (SERCA2a). This protein has an important role in removing cytoplasmic Ca++ and is the main mechanism for restoring SR Ca++ load during the contraction cycle.
- The transcription factor STAT5 requires O-GlcNAc to bind to the coactivator of transcription (CBP), which is an essential interaction for STAT5-mediated gene transcriptions. Other members of the STAT family are also O-GlcNAcylated, but the function of O-GlcNAc on these proteins is unknown.
- Increased O-GlcNAcylation of PDX-1, a pancreatic β-cell transcription factor that controls insulin transcription, increases its affinity for DNA and results in increased proinsulin transcription.
- O-GlcNAcylation prevents YY1 (a zinc-finger, DNA-binding transcription factor that regulates the expression of a wide variety of cellular and viral genes and is essential for the development of mammalian embryos) from binding the retinoblastoma Rb protein. Upon dissociation from Rb, the O-GlcNAcylated YY1 is free to bind DNA and activate transcription.
- The Rb tumor suppressor protein is itself O-GlcNAcylated during the stages of the cell cycle when it is not O-phosphorylated. It is the O-GlcNAcylated form of Rb that binds to E2F transcription factors.
- Elf-1 (a member of the Ets transcription factor family) is involved in the transcriptional regulation of several hematopoietic genes. Following O-GlcNAcylation and phosphorylation by PKC-θ, the cytoplasm-located Elf-1 translocates to the nucleus where it binds to the promoter of the T-cell receptor (TCR) ζ gene and promotes its transcription.
- O-GlcNAcylation of CREB (cyclic-AMP-responsive element-binding protein), a transcription factor essential for long-term memory, disrupts the interaction between CREB and component of the basal transcription machinery, thereby TAFII130, a repressing the transcriptional activity of CREB.
- O-GlcNAcylation of Sp1 and estrogen receptors increases the steady-state levels of these transcription factors by preventing their degradation.
Thus, it appears that the glycan is important in regulating the complex assembly and protein interactions required to control both the specificity and activity of the transcriptional machinery. At least 15 well-characterized ribosomal proteins and several associated translational factors are O-GlcNAcylated. It has been proposed that the activity of eukaryotic initiation factor 2 (eIF2) is regulated by its binding to p67. The p67 protein is O-GlcNAcylated, and its interactions with eIF2 are controlled by the O-GlcNAc modification. Thus, although little work has been done with respect to O-GlcNAcylation and the regulation of protein translation, it is likely that O-GlcNAc is also involved in these processes.
O-GlcNAc Regulates Protein Trafficking and Turnover
In neurons, O-GlcNAcylation of synapsin proteins appears to prevent release of synaptic vesicles from the cytoskeleton, thus regulating the release of neurotransmitters at the synapse. O-GlcNAcylation also appears to regulate the trafficking of β-catenin and E-cadherin to the cell surface of epithelial cells. When these proteins are O-GlcNAcylated, they remain bound to the cytoskeleton and are not transported to the cell surface. Because E-cadherin is an important cell adhesion molecule, blockage of its transport to the cell surface may be important to epithelial tumor cell metastasis. Likewise, there is growing evidence that increased O-GlcNAcylation of GLUT4 vesicle proteins such as Munc18 and others has a role in the inhibition of glucose transport in diabetes. GLUT4 is the insulin-sensitive glucose transporter that, when stimulated by insulin in muscle and adipose tissues, is rapidly transported to the plasma membrane to increase glucose uptake.
As mentioned above, several studies have shown that O-GlcNAcylation of proteins, either directly or indirectly, slows their proteolytic degradation. The 26S proteasome is a complex structure that degrades many ubiquitin-tagged regulatory proteins within the cell. Recent proteomic analyses of the 26S proteasome have demonstrated that 5 of 19 and 9 of 14 proteins of the catalytic and regulatory cores, respectively, are modified by O-GlcNAc. Increased O-GlcNAcylation of the Rpt2 ATPase, a component of the 19S cap of the proteasome, blocks its ATPase activity, reducing proteasome-catalyzed degradation. It has been suggested that O-GlcNAcylation of the proteasome allows the cell to respond to metabolic needs by controlling the availability of amino acids and altering the half-lives of key regulatory proteins.
O-GlcNAc Is Involved in Neurodegenerative Disease
The OGT gene has been mapped to Xq13, a locus linked to several neurological diseases including Parkinson’s dystonia. Similarly, as noted above, the gene encoding O-GlcNAcase is located at 10q24.1, a locus linked to late-onset Alzheimer’s disease (AD). Brain metabolic processes, including glycolysis, and overall levels of glucose are known to decline with age, particularly in AD neurons. As discussed above, one normal function of O-GlcNAc is to “cap” phosphorylation sites to shut them off. Most of the proteins involved in the pathology of AD are modified by O-GlcNAc. Tau, a microtubule-associated protein that has multiple microtubule-binding repeats, is required for polymerization and stability of microtubules in neurons. There are numerous kinases known to phosphorylate tau including glycogen synthase kinase 3 (GSK-3), CKII, and extracellular regulated protein kinase (ERK). Hyperphosphorylated tau aggregates to form neurofibrillary tangles (PHF-tau) in AD neurons. Normally, in adult brain, tau protein is extensively modified by O-GlcNAc at more than 12 sites. At least one identified O-GlcNAc site in tau is in its microtubule-binding domain. This site (Ser-262) accounts for 70% of PHF-tau-forming activity in vitro. It is postulated that O-GlcNAc modification of tau may affect its microtubule-binding ability, compete with phosphorylation, or even affect its nucleocytoplasmic localization. Recent studies on human brain tau have established that O-GlcNAcylation negatively regulates its O-phosphorylation in a site-specific manner both in vitro and in vivo. Using an animal model of starved mice, low glucose uptake/metabolism mimicked the hypo-O-GlcNAcylation and hyper-O-phosphorylation of tau seen in human AD brain. Quantitative comparisons between normal human brain tissue and that of AD patients indeed showed that global O-GlcNAcylation is reduced in AD brains.
The β-amyloid precursor protein (APP) is O-GlcNAc modified in its cytoplasmic tail; however, the sites of modification are not known. APP is also phosphorylated in its cytoplasmic tail by cyclin-dependent kinase 1, which is known to affect its proteolytic processing. Upon abnormal proteolysis, APP gives rise to the toxic β1-42 peptide fragment, which forms the amyloid plaques present in AD brains. The cytoplasmic tail also contains a sequence controlling endocytosis and a G-protein-binding site, suggesting that O-GlcNAcylation of this region of APP may affect signaling events, trafficking, and metabolism of APP.
Altered O-GlcNAcylation has been reported for other proteins involved in neurodegenerative disease. Neurofilaments appear to be hypo-O-GlcNAcylated in neurons from patients with Lou Gehrig’s disease (ALS; amyotrophic lateral sclerosis). Clathrin-assembly proteins AP-3 and AP-180 are both modified by O-GlcNAc, and these modifications decline in AD, suggesting that reduced O-GlcNAc is associated with the loss of synaptic vesicle recycling. Altogether, current data point to potentially significant roles of the O-GlcNAc modification in normal neuronal function and in the molecular mechanisms underlying the pathology of neurodegenerative disease.
Elevated O-GlcNAc Underlies Diabetes and Glucose Toxicity
Perhaps the best understood function of O-GlcNAc is its role in the regulation of insulin signaling and as a mediator of glucose toxicity. Increased O-GlcNAc in adipocytes or muscle blocks insulin signaling at several points (Figure 18.8), and overexpression of OGT in muscle or adipose tissue in transgenic mice causes overt diabetes. Many of the toxic effects of glucose require its conversion to glucosamine, which concomitantly elevates UDP-GlcNAc and increases O-GlcNAcylation. Current models suggest that abnormal increases in O-GlcNAcylation, due to hyperglycemia, hyperlipidemia, and/or hyperinsulinemia, disturb the normal dynamic balance between O-GlcNAcylation and O-phosphorylation that controls signaling, transcription, and other cellular functions, leading to the toxicity associated with diabetes.

FIGURE 18.8
Elevating O-GlcNAc blocks insulin signaling at many points. Glucose flux via glucose transporters (e.g., GLUT4 in insulin-sensitive cells) through the hexosamine biosynthetic pathway (which accounts for 2–5% of total glucose utilization) leads (more...)
The first studies to link glucosamine metabolism directly with the toxicity of glucose in diabetes showed that glucosamine is many times more potent than glucose in inducing insulin resistance in cultured adipocytes. These studies also demonstrated that the ability of glucose to induce insulin resistance could be blocked by deoxynorleucine (DON), a drug that inhibits glucose:fructose amidotransferase (GFAT, the enzyme that converts fructose-6-P to glucosamine-6-P; Figure 18.8), and that this blockage could be bypassed by adding glucosamine to the culture media (see Chapter 4). Insulin resistance is the hallmark of type II diabetes. Later studies found that increased glucosamine availability induced severe skeletal muscle insulin resistance in normal rats and also showed that glucosamine metabolism has a role in fat-induced insulin resistance in vivo. Elevated fatty acids, like hyperglycemia, induce insulin resistance by increasing production of glucosamine metabolites. O-GlcNAcylation of glycogen synthase prevents its activation by insulin, blocking the storage of glucose. A recent genetic study showed a strong association between a single nucleotide polymorphism in O-GlcNAcase and a population of individuals with type II diabetes. In rat skeletal muscle, hyperinsulinemia, as seen in type II diabetes, dramatically increases the levels of O-GlcNAc–modified proteins. Infusion of glucosamine also increases the levels of O-GlcNAc–modified proteins in muscle, even in the absence of elevated insulin.
Many studies have shown that hyperglycemia and/or hyperinsulinemia globally increases the O-GlcNAcylation of proteins in different cell types. Several studies have shown that elevated glucose increases the O-GlcNAcylation of proteins in pancreatic β-cells (insulin-secreting cells), and as described above, O-GlcNAcylation of transcription factors may regulate insulin gene transcription. A single dose of a chemically reactive analog of GlcNAc (streptozotocin, STZ) given to animals selectively kills the β-cells of their pancreas, resulting in type I diabetes. The highly selective nature of STZ suggests that β-cells are particularly sensitive to altered glucosamine metabolism. Biochemical and histological studies of rat aorta and cultured rat aortic smooth muscle show that hyperglycemia qualitatively and quantitatively alters the expression of many O-GlcNAcylated proteins, suggesting a role of O-GlcNAc in glucose toxicity to vascular tissues. Transgenic mice that overexpress the glucose transporter protein (GLUT1) in muscle have increased UDP-GlcNAc levels, are insulin resistant, and have increased O-GlcNAcylation of GLUT4-vesicle-associated proteins. Free fatty acids increase UDP-GlcNAc in cultured human myotubes and increase the O-GlcNAc-dependent DNA-binding activity of the Sp1 transcription factor. Studies of cultured myotubes grown in high glucose and/or insulin showed enhanced O-GlcNAcylation of numerous proteins. Proteomic analyses of these myotubes show that hyper-O-GlcNAcylated proteins include HSP70, α-tubulin, and Sp1. The roles of O-GlcNAc in diabetic cardiomyopathy have been examined in a rat model. Myocytes exposed to high glucose display altered O-GlcNAcylation of several transcription factors involved in cardiomyocyte function, concomitant with altered calcium signaling. In contrast, when the cardiomyocytes are transfected with adenovirus encoding O-GlcNAcase, cardiomyocyte functions improve dramatically. Thus, it is now clear that both hyperglycemia and insulin regulate the O-GlcNAcylation state of myriad cellular proteins, which in turn modulates signaling and transcription. Although much remains to be learned, it is increasingly clear that dysregulation of O-GlcNAcylation underlies the molecular basis of glucose toxicity and insulin resistance, two major hallmarks of diabetes.
O-GlcNAc and Stress Survival
In every mammalian cell type examined to date, one of the earliest responses to cellular stress is a rapid and global increase in O-GlcNAcylation on a multitude of proteins. Levels of O-GlcNAc increase very rapidly in response to every form of stress (heat shock, ethanol, UV, hypoxia, reductive, oxidative, and osmotic stress). Moreover, the stress-induced increase in O-GlcNAcylation of many proteins is dynamic, returning to normal after 24–48 hours. This increase in O-GlcNAc could be a result of increased glucose flux into the cells that occurs in response to stress, or increased activity of OGT, decreased activity of O-GlcNAcase, or all three combined.
Artificial modulation of the levels of O-GlcNAc results in altered tolerance to lethal levels of cellular stress. Decreasing OGT and O-GlcNAc levels results in cells that are less tolerant, whereas increasing the levels results in cells that are more tolerant. Short-term elevation in O-GlcNAcylation appears to be an important survival mechanism. The overexpression of heat-shock proteins (chaperones) is also known to protect cells from stress. The induction of heat-shock proteins (HSP70 and HSP40) occurs faster, and these proteins turn over more slowly in the presence of increased O-GlcNAc. Furthermore, reduced levels of HSP70 and HSP40 are observed in cell lines in which OGT is reduced. It is clear that O-GlcNAc mediates stress tolerance, in part, by altering the levels of heat-shock proteins. However, O-GlcNAc may also protect cells in other ways, for example, by (1) stabilizing protein structure; (2) preventing protein–protein aggregation, possibly directly or through HSP70’s O-GlcNAc-binding activity; (3) modulating signal transduction pathways implicated previously in glucose protection, such as JNK activation; and finally (4) altering the activity or localization of heat-shock proteins, many of which are known to be modified by O-GlcNAc. Regardless of the underlying mechanisms, it is clear that increased O-GlcNAcylation on many proteins is one of the most rapid cellular responses to stress from a wide variety of unrelated sources. Elucidating how increasing O-GlcNAcylation helps a cell to survive stressful conditions should improve our understanding of the roles of this ubiquitous posttranslational modification.
FURTHER READING
- Torres C.-R, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed: 6421821]
- Hart GW. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu Rev Biochem. 1997;66:315–335. [PubMed: 9242909]
- Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem. 2000;275:29179–29182. [PubMed: 10924527]
- Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. [PubMed: 11269319]
- Vocadlo DJ, Hang HC, Kim E.-J, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci. 2003;100:9116–9121. [PMC free article: PMC171382] [PubMed: 12874386]
- Wells L, Vosseller K, Hart GW. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell Mol Life Sci. 2003;60:222–228. [PubMed: 12678487]
- Whelan SA, Hart GW. Proteomic approaches to analyze the dynamic relationships between nucleocytoplasmic protein glycosylation and phosphorylation. Circ Res. 2003;93:1047–1058. [PubMed: 14645135]
- Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci. 2004;101:13132–13137. [PMC free article: PMC516536] [PubMed: 15340146]
- Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong C.-X. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci. 2004;101:10804–10809. [PMC free article: PMC490015] [PubMed: 15249677]
- Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta. 2004;1673:13–28. [PubMed: 15238246]
- Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code” Science STKE. 2005;312:re13. [PubMed: 16317114]
- Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: How a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem. 2006;97:71–83. [PubMed: 16237703]
- Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. [PubMed: 17460662]
- Review The O-GlcNAc Modification.[Essentials of Glycobiology. 2015]Review The O-GlcNAc Modification.Zachara N, Akimoto Y, Hart GW. Essentials of Glycobiology. 2015
- Review The O-GlcNAc Modification.[Essentials of Glycobiology. 2022]Review The O-GlcNAc Modification.Zachara NE, Akimoto Y, Boyce M, Hart GW. Essentials of Glycobiology. 2022
- Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine.[Anal Biochem. 2001]Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Anal Biochem. 2001 Jun 15; 293(2):169-77.
- Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry.[Anal Biochem. 1996]Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry.Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, Hart GW. Anal Biochem. 1996 Feb 1; 234(1):38-49.
- Role of O-linked beta-N-acetylglucosamine modification in the subcellular distribution of alpha4 phosphoprotein and Sp1 in rat lymphoma cells.[J Cell Biochem. 2005]Role of O-linked beta-N-acetylglucosamine modification in the subcellular distribution of alpha4 phosphoprotein and Sp1 in rat lymphoma cells.Dauphinee SM, Ma M, Too CK. J Cell Biochem. 2005 Oct 15; 96(3):579-88.
- The O-GlcNAc Modification - Essentials of GlycobiologyThe O-GlcNAc Modification - Essentials of Glycobiology
Your browsing activity is empty.
Activity recording is turned off.
See more...

