NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Lizarraga LE, Patlewicz G, Dean JL II, et al. Provisional Peer-Reviewed Toxicity Values for 2,3-Toluenediamine (CASRN 2687-25-4). Cincinnati (OH): U.S. Environmental Protection Agency; 2021 Aug.

Provisional Peer-Reviewed Toxicity Values for 2,3-Toluenediamine (CASRN 2687-25-4).
Show detailsFor reasons noted in the main Provisional Peer-Reviewed Toxicity Value (PPRTV) document, there is inadequate information to assess the carcinogenic potential of 2,3-toluenediamine (2,3-TDA). However, information is available for this chemical which, although insufficient to support a weight-of-evidence (WOE) descriptor and derivation of provisional cancer risk estimates under current guidelines, may be of use to risk assessors. In such cases, the Center for Public Health and Environmental Assessment (CPHEA) summarizes available information in an appendix and develops a “screening evaluation of potential carcinogenicity.” Appendices receive the same level of internal and external scientific peer review as the provisional cancer assessments in PPRTVs to ensure their appropriateness within the limitations detailed in the document. Users of the information regarding potential carcinogenicity in this appendix should understand that there could be more uncertainty associated with this evaluation than for the cancer WOE descriptors presented in the body of the assessment. Questions or concerns about the appropriate use of the screening evaluation of potential carcinogenicity should be directed to the CPHEA.
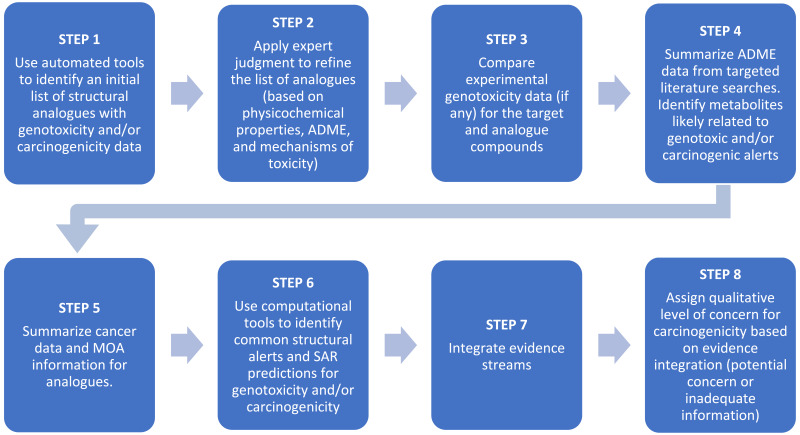
The screening evaluation of potential carcinogenicity includes the general steps shown in Figure B-1. The methods for Steps 1–8 apply to any target chemical and are described in this appendix. Chemical-specific data for all steps in this process are summarized in Appendix C.
STEP 1. USE OF AUTOMATED TOOLS TO IDENTIFY STRUCTURAL ANALOGUES WITH CARCINOGENICITY AND/OR GENOTOXICITY DATA
ChemACE Clustering
The U.S. EPA’s Chemical Assessment Clustering Engine [ChemACE; U.S. EPA (2011a)] is an automated tool that groups (or clusters) a user-defined list of chemicals based on chemical structure fragments. The methodology used to develop ChemACE was derived from U.S. EPA’s Analog Identification Methodology (AIM) tool, which identifies structural analogues for a chemical based on common structural fragments. ChemACE uses the AIM structural fragment recognition approach for analogue identification and applies advanced queries and user-defined rules to create the chemical clusters. The ChemACE cluster outputs are available in several formats and layouts (i.e., Microsoft Excel, Adobe PDF) to allow rapid evaluation of structures, properties, mechanisms, and other parameters which are customizable based on an individual user’s needs. ChemACE clustering has been successfully used with chemical inventories for identifying trends within a series of structurally similar chemicals, demonstrating structural diversity in a chemical inventory, and detecting structural analogues to fill data gaps and/or perform read-across analysis.
For this project, ChemACE is used to identify potential structural analogues of the target compound that have available carcinogenicity assessments and/or carcinogenicity data. An overview of the ChemACE process in shown in Figure B-2.
The chemical inventory was populated with chemicals from the following databases and lists:
- Carcinogenic Potency Database [CPDB; CPDB (2011)]
- Agents classified by the International Agency for Research on Cancer (IARC) monographs (IARC, 2018)
- National Toxicology Program (NTP) Report on Carcinogens [ROC; NTP (2016a)]
- NTP technical reports (NTP, 2017)
- Integrated Risk Information System (IRIS) carcinogens (U.S. EPA, 2017)
- California EPA (CalEPA) Prop 65 list (CalEPA, 2017)
- European Chemicals Agency (ECHA) carcinogenicity data available in the Organisation for Economic Co-operation and Development (OECD) Quantitative Structure-Activity Relationship (QSAR) Toolbox (OECD, 2017)
- PPRTVs for Superfund (U.S. EPA, 2020b)
In total, 2,123 distinct substances were identified from the sources above. For the purpose of ChemACE clustering, each individual substance needed to meet the following criteria:
- 1)
Substance is not a polymer, metal, inorganic, or complex salt because ChemACE is not designed to accommodate these substances;
- 2)
Substance has CASRN or unambiguous chemical identification; and
- 3)
A unique Simplified Molecular Input Line Entry System (SMILES) notation (encoded molecular structure format used in ChemACE) for the substance can be identified from one of these sources:
- Syracuse Research Corporation (SRC) and Distributed Structure-Searchable Toxicity (DSSTox) lists of known SMILES associated with unique CASRNs (the combined lists contained >200,000 SMILES); or
- ChemIDplus, U.S. EPA CompTox Chemicals Dashboard, or internet searches.
Of the initial list of 2,123 substances, 201 were removed because they did not meet one of the first two criteria, and 155 were removed because they did not meet the third. The final inventory of substances contained 1,767 unique compounds.
Two separate ChemACE approaches were compared for clustering of the chemical inventory. The restrictive clustering approach, in which all compounds in a cluster contain all of the same fragments and no different fragments, resulted in 208 clusters. The less restrictive approach included the following rules for remapping the chemical inventory:
- treat adjacent halogens as equivalent, allowing fluorine (F) to be substituted for chlorine (Cl), Cl for bromine (Br), Br for iodine (I);
- allow methyl, methylene, and methane to be equivalent;
- allow primary, secondary, and tertiary amines to be equivalent; and
- exclude aromatic thiols (removes thiols from consideration).
Clustering using the less restrictive approach (Pass 2) resulted in 284 clusters. ChemACE results for clustering of the target chemical within the clusters of the chemical inventory are described in Appendix C.
Analogue Searches in the OECD QSAR Toolbox (Dice Method)
The OECD QSAR Toolbox (Version 4.1) is used to search for additional structural analogues of the target compound. There are several structural similarity score equations available in the Toolbox (Dice, Tanimoto, Kulczynski-2, Ochiai/Cosine, and Yule). Dice is considered the default equation. The specific options that are selected for the performance of this search include a comparison of molecular features (atom-centered fragments) and atom characteristics (atom type, count hydrogens attached, and hybridization). Chemicals identified in these similarity searches are selected if their similarity scores exceeded 50%.
The OECD QSAR Toolbox Profiler is used to identify those structural analogues from the Dice search that have carcinogenicity and/or genotoxicity data. Nine databases in the OECD QSAR Toolbox (Version 4.1) provide data for carcinogenicity or genotoxicity (see Table B-1).
Analogue search results for the target chemical are described in Appendix C.
Table B-1Databases Providing Carcinogenicity and Genotoxicity Data in the OECD QSAR Toolbox (Version 4.1)
| Database Name | Toolbox Database Descriptiona |
|---|---|
| CPDB | The CPDB provides access to bioassay literature with qualitative and quantitative analysis of published experiments from the general literature (through 2001) and from the NCI/NTP (through 2004). Reported results include bioassays in rats, mice, hamsters, dogs, and nonhuman primates. A calculated carcinogenic potency (TD50) is provided to standardize quantitative measures for comparison across chemicals. The CPDB contains 1,531 chemicals and 3,501 data points. |
| ISSCAN | The ISSCAN database provides information on carcinogenicity bioassays in rats and mice reported in sources including NTP, CPDB, CCRIS, and IARC. This database reports a carcinogenicity TD50. There are 1,149 chemicals and 4,518 data points included in the ISSCAN database. |
| ECHA CHEM | The ECHA CHEM database provides information on chemicals manufactured or imported in Europe from registration dossiers submitted by companies to ECHA to comply with the REACH Regulation framework. The ECHA database includes 9,229 chemicals with almost 430,000 data points for a variety of endpoints including carcinogenicity and genotoxicity. ECHA does not verify the information provided by the submitters. |
| ECVAM Genotoxicity and Carcinogenicity | The ECVAM Genotoxicity and Carcinogenicity database provides genotoxicity and carcinogenicity data for Ames positive chemicals in a harmonized format. ECVAM contains in vitro and in vivo bacteria mutagenicity, carcinogenicity, CA, CA/aneuploidy, DNA damage, DNA damage and repair, mammalian culture cell mutagenicity, and rodent gene mutation data for 744 chemicals and 9,186 data points. |
| ISSCTA | ISSCTA provides results of four types of in vitro cell transformation assays including Syrian hamster embryo cells, mouse BALB/c 3T3, mouse C3H/10T1/2, and mouse Bhas 42 assays that inform nongenotoxic carcinogenicity. ISSCTA consists of 352 chemicals and 760 data points. |
| Bacterial mutagenicity ISSSTY | The ISSSTY database provides data on in vitro Salmonella typhimurium Ames test mutagenicity (positive and negative) taken from the CCRIS database in TOXNET. The ISSSTY database provides data for 7,367 chemicals and 41,634 data points. |
| Genotoxicity OASIS | The Genotoxicity OASIS database provides experimental results for mutagenicity results from “Ames tests (with and without metabolic activation), in vitro chromosomal aberrations and MN and MLA evaluated in vivo and in vitro, respectively.” The Genotoxicity OASIS database consists of 7,920 chemicals with 29,940 data points from 7 sources. |
| Micronucleus OASIS | The Micronucleus OASIS database provides experimental results for in vivo bone marrow and peripheral blood MNT CA studies in blood erythrocytes, bone marrow cells, and polychromatic erythrocytes of humans, mice, rabbits, and rats for 557 chemicals. |
| ISSMIC | The ISSMIC database provides data on the results of in vivo MN mutagenicity assay to detect CAs in bone marrow cells, peripheral blood cells, and splenocytes in mice and rats. Sources include TOXNET, NTP, and the Leadscope FDA CRADA Toxicity Database. The ISSMIC database includes data for 563 chemicals and 1,022 data points. |
- a
Descriptions were obtained from the OECD QSAR Toolbox documentation [Version 4.1; OECD (2017)].
CA = chromosomal aberration; CCRIS = Chemical Carcinogenesis Research Information System; CPBD = Carcinogenic Potency Database; CRADA = cooperative research and development agreement; DNA = deoxyribonucleic acid; ECHA = European Chemicals Agency; ECVAM = European Centre for the Validation of Alternative Methods; FDA = Food and Drug Administration; IARC = International Agency for Research on Cancer; ISSCAN = Istituto Superiore di Sanità Chemical Carcinogen; ISSCTA = Istituto Superiore di Sanità Cell Transformation Assay; ISSMIC = Istituto Superiore di Sanità Micronucleus; ISSSTY = Istituto Superiore di Sanità Salmonella typhimurium; MLA = mouse lymphoma gene mutation assay; MN = micronuclei; MNT = micronucleus test; NCI = National Cancer Institute; NTP = National Toxicology Program; OECD = Organization for Economic Co-operation and Development; QSAR = quantitative structure-activity relationship; REACH = Registration, Evaluation, Authorization and Restriction of Chemicals; TD50 = median toxic dose.
STEPS 2–5. ANALOGUE REFINEMENT AND SUMMARY OF EXPERIMENTAL DATA FOR GENOTOXICITY, TOXICOKINETICS, CARCINOGENICITY, AND MODE OF ACTION
The outcome of the Step 1 analogue identification process using ChemACE and the OECD QSAR Toolbox is an initial list of structural analogues with genotoxicity and/or carcinogenicity data. Expert judgment is applied in Step 2 to refine the list of analogues based on physicochemical properties; absorption, distribution, metabolism, and excretion (ADME); and mechanisms of toxicity. The analogue refinement process is chemical-specific and is described in Appendix C. Steps 3, 4, and 5 (summary of experimental data for genotoxicity, toxicokinetics, carcinogenicity, and mode of action [MOA]) are also chemical specific (see Appendix C for further details).
STEP 6. STRUCTURAL ALERTS AND STRUCTURE-ACTIVITY RELATIONSHIP PREDICTIONS FOR 2,3-TDA AND ANALOGUES
Structural alerts (SAs) and predictions for genotoxicity and carcinogenicity are identified using six freely available structure-based tools (described in Table B-2).
Table B-2Tools Used to Identify SAs and Predict Carcinogenicity and Genotoxicity
| Name | Descriptiona |
|---|---|
| OECD QSAR Toolbox (Version 4.1) | Seven OECD QSAR Toolbox profiling methods were used, including:
|
| OncoLogic (Version 7) | OncoLogic is a tool for predicting the potential carcinogenicity of chemicals based on the application of rules for SAR analysis, developed by experts. Results may range from “low” to “high” concern level. |
| ToxAlerts | ToxAlerts is a platform for screening chemical compounds against SAs, developed as an extension to the OCHEM system (https://ochem
|
| ToxRead (Version 0.9) | ToxRead is a tool designed to assist in making read-across evaluations reproducible. SAs for mutagenicity are extracted from similar molecules with available experimental data in its database. Five similar compounds were selected for this project. The rule sets included:
|
| Toxtree (Version 2.6.13) | Toxtree estimates toxic hazard by applying a decision tree approach. Chemicals were queried in Toxtree using the Benigni/Bossa rulebase for mutagenicity and carcinogenicity. If a potential carcinogenic alert based on any QSAR model or if any SA for genotoxic and nongenotoxic carcinogenicity was reported, then the prediction was recorded as a positive carcinogenicity prediction for the test chemical. The output definitions from the tool manual are listed below:
|
| VEGA | VEGA applies several QSARs to a given chemical, as described below:
|
- a
There is some overlap between the tools. For example, OncoLogic classification is provided by the QSAR Toolbox, but the prediction is available only through OncoLogic, and alerts or decision trees were used in or adapted from several models (e.g., Benigni and Bossa alerts and Toxtree decision tree) (OECD, 2017).
ANTARES = Alternative Non-Testing Methods Assessed for REACH Substances; CA = chromosomal aberration; CAESAR = Computer Assisted Evaluation of industrial chemical Substances According to Regulations; CONSENSUS = consensus assessment based on multiple models (CAESAR, SARpy, ISS, and k-NN); CRS4 = Center for Advanced Studies, Research and Development in Sardinia; CPDB = Carcinogenic Potency Database; DNA = deoxyribonucleic acid; FN = false negative; IRFMN = Istituto di Ricerche Farmacologiche Mario Negri; ISS = Istituto Superiore di Sanità; ISSCAN-CGX = Istituto Superiore di Sanità Chemical Carcinogen; k-NN = k-nearest neighbor; LMC = Laboratory for Mathematical Chemistry; MN = micronucleus; MNT = micronucleus test; OCHEM = Online Chemical Monitoring Environment; OECD = Organisation for Economic Co-operation and Development; QSAR = quantitative structure-activity relationship; REACH = Registration, Evaluation, Authorisation and Restriction of Chemicals; SA = structural alert; SAR = structure-activity relationship; SVM = support vector machine; TIMES = The Integrated MARKEL-EFOM System; VEGA = Virtual models for property Evaluation of chemicals within a Global Architecture.
The tool results for the target and analogue compounds are provided in Appendix C.
STEP 7. EVIDENCE INTEGRATION FOR SCREENING EVALUATION OF 2,3-TDA CARCINOGENICITY
Data identified across multiple lines of evidence from Steps 1–6 (outlined above) are integrated to determine the qualitative level of concern for potential carcinogenicity of the target compound (Step 8). In the absence of information supporting carcinogenic portal-of-entry effects, the qualitative level of concern for the target chemical should be considered applicable to all routes of exposure.
Evidence integration for the target compound is provided in Appendix C.
- USE OF AUTOMATED TOOLS TO IDENTIFY STRUCTURAL ANALOGUES WITH CARCINOGENICITY AND/OR GENOTOXICITY DATA
- ANALOGUE REFINEMENT AND SUMMARY OF EXPERIMENTAL DATA FOR GENOTOXICITY, TOXICOKINETICS, CARCINOGENICITY, AND MODE OF ACTION
- STRUCTURAL ALERTS AND STRUCTURE-ACTIVITY RELATIONSHIP PREDICTIONS FOR 2,3-TDA AND ANALOGUES
- EVIDENCE INTEGRATION FOR SCREENING EVALUATION OF 2,3-TDA CARCINOGENICITY
- BACKGROUND AND METHODOLOGY FOR THE SCREENING EVALUATION OF POTENTIAL CARCINOGENI...BACKGROUND AND METHODOLOGY FOR THE SCREENING EVALUATION OF POTENTIAL CARCINOGENICITY - Provisional Peer-Reviewed Toxicity Values for 2,3-Toluenediamine (CASRN 2687-25-4)
Your browsing activity is empty.
Activity recording is turned off.
See more...