By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002.

Molecular Biology of the Cell. 4th edition.
Show detailsAll eucaryotic cells have an endoplasmic reticulum (ER). Its membrane typically constitutes more than half of the total membrane of an average animal cell (see Table 12-2). The ER is organized into a netlike labyrinth of branching tubules and flattened sacs extending throughout the cytosol (Figure 12-35). The tubules and sacs are all thought to interconnect, so that the ER membrane forms a continuous sheet enclosing a single internal space. This highly convoluted space is called the ER lumen or the ER cisternal space, and it often occupies more than 10% of the total cell volume (see Table 12-1). The ER membrane separates the ER lumen from the cytosol, and it mediates the selective transfer of molecules between these two compartments.

Figure 12-35
Fluorescent micrographs of the endoplasmic reticulum. (A) Part of the ER network in a cultured mammalian cell, stained with an antibody that binds to a protein retained in the ER. The ER extends as a network throughout the entire cytosol, so that all (more...)

The ER has a central role in lipid and protein biosynthesis. Its membrane is the site of production of all the transmembrane proteins and lipids for most of the cell's organelles, including the ER itself, the Golgi apparatus, lysosomes, endosomes, secretory vesicles, and the plasma membrane. The ER membrane makes a major contribution to mitochondrial and peroxisomal membranes by producing most of their lipids. In addition, almost all of the proteins that will be secreted to the cell exterior—plus those destined for the lumen of the ER, Golgi apparatus, or lysosomes—are initially delivered to the ER lumen.
Membrane-bound Ribosomes Define the Rough ER
The ER captures selected proteins from the cytosol as they are being synthesized. These proteins are of two types: transmembrane proteins, which are only partly translocated across the ER membrane and become embedded in it, and water-soluble proteins, which are fully translocated across the ER membrane and are released into the ER lumen. Some of the transmembrane proteins function in the ER, but many are destined to reside in the plasma membrane or the membrane of another organelle. The water-soluble proteins are destined either for the lumen of an organelle or for secretion. All of these proteins, regardless of their subsequent fate, are directed to the ER membrane by the same kind of signal sequence and are translocated across it by similar mechanisms.
In mammalian cells, the import of proteins into the ER begins before the polypeptide chain is completely synthesized—that is, import is a co-translational process. This distinguishes the process from the import of proteins into mitochondria, chloroplasts, nuclei, and peroxisomes, which are posttranslational processes. Since one end of the protein is usually translocated into the ER as the rest of the polypeptide chain is being made, the protein is never released into the cytosol and therefore is never in danger of folding up before reaching the translocator in the ER membrane. Thus, in contrast to the posttranslational import of proteins into mitochondria and chloroplasts, chaperone proteins are not required to keep the protein unfolded. The ribosome that is synthesizing the protein is directly attached to the ER membrane. These membrane-bound ribosomes coat the surface of the ER, creating regions termed rough endoplasmic reticulum, or rough ER (Figure 12-36A).

Figure 12-36
The rough ER. (A) An electron micrograph of the rough ER in a pancreatic exocrine cell that makes and secretes large amounts of digestive enzymes every day. The cytosol is filled with closely packed sheets of ER membrane studded with ribosomes. At the (more...)
There are therefore two spatially separate populations of ribosomes in the cytosol. Membrane-bound ribosomes, attached to the cytosolic side of the ER membrane, are engaged in the synthesis of proteins that are being concurrently translocated into the ER. Free ribosomes, unattached to any membrane, synthesize all other proteins encoded by the nuclear genome. Membrane-bound and free ribosomes are structurally and functionally identical. They differ only in the proteins they are making at any given time. When a ribosome happens to be making a protein with an ER signal sequence, the signal directs the ribosome to the ER membrane.
Since many ribosomes can bind to a single mRNA molecule, a polyribosome is usually formed, which becomes attached to the ER membrane, directed there by the signal sequences on multiple growing polypeptide chains (Figure 12-36B). The individual ribosomes associated with such an mRNA molecule can return to the cytosol when they finish translation near the 3′ end of the mRNA molecule. The mRNA itself, however, remains attached to the ER membrane by a changing population of ribosomes, each transiently held at the membrane by the translocator. In contrast, if an mRNA molecule encodes a protein that lacks an ER signal sequence, the polyribosome that forms remains free in the cytosol, and its protein product is discharged there. Therefore, only those mRNA molecules that encode proteins with an ER signal sequence bind to rough ER membranes; those mRNA molecules that encode all other proteins remain free in the cytosol. Individual ribosomal subunits are thought to move randomly between these two segregated populations of mRNA molecules (Figure 12-37).
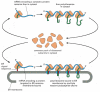
Figure 12-37
Free and membrane-bound ribosomes. A common pool of ribosomes is used to synthesize the proteins that stay in the cytosol and those that are transported into the ER. The ER signal sequence on a newly formed polypeptide chain directs the engaged ribosome (more...)
Smooth ER Is Abundant in Some Specialized Cells
Regions of ER that lack bound ribosomes are called smooth endoplasmic reticulum, or smooth ER. In the great majority of cells, such regions are scanty and are often partly smooth and partly rough. They are sometimes called transitional ER because they contain ER exit sites from which transport vesicles carrying newly synthesized proteins and lipids bud off for transport to the Golgi apparatus. In certain specialized cells, however, the smooth ER is abundant and has additional functions. In particular, it is usually prominent in cells that specialize in lipid metabolism. Cells that synthesize steroid hormones from cholesterol, for example, have an expanded smooth ER compartment to accommodate the enzymes needed to make cholesterol and to modify it to form the hormones (Figure 12-38A).

Figure 12-38
The smooth ER. (A) Abundant smooth ER in a steroid-hormone-secreting cell. This electron micrograph is of a testosterone-secreting Leydig cell in the human testis. (B) A three-dimensional reconstruction of a region of smooth ER and rough ER in a liver (more...)
The main cell type in the liver, the hepatocyte, is another cell with an abundant smooth ER. It is the principal site of production of lipoprotein particles, which carry lipids via the bloodstream to other parts of the body. The enzymes that synthesize the lipid components of lipoproteins are located in the membrane of the smooth ER, which also contains enzymes that catalyze a series of reactions to detoxify both lipid-soluble drugs and various harmful compounds produced by metabolism. The most extensively studied of these detoxification reactions are carried out by the cytochrome P450 family of enzymes, which catalyze a series of reactions in which water-insoluble drugs or metabolites that would otherwise accumulate to toxic levels in cell membranes are rendered sufficiently water-soluble to leave the cell and be excreted in the urine. Because the rough ER alone cannot house enough of these and other necessary enzymes, a major portion of the membrane in a hepatocyte normally consists of smooth ER (Figure 12-38B; see Table 12-2).
When large quantities of certain compounds, such as the drug phenobarbital, enter the circulation, detoxification enzymes are synthesized in the liver in unusually large amounts, and the smooth ER doubles in surface area within a few days. Once the drug has disappeared, the excess smooth ER membrane is specifically and rapidly removed by a lysosome-dependent process called autophagocytosis (discussed in Chapter 13). It is not known how these dramatic changes are regulated.
Another function of the ER in most eucaryotic cells is to sequester Ca2+ from the cytosol. The release of Ca2+ into the cytosol from the ER, and its subsequent reuptake, is involved in many rapid responses to extracellular signals, as discussed in Chapter 15. The storage of Ca2+ in the ER lumen is facilitated by the high concentrations of Ca2+-binding proteins there. In some cell types, and perhaps in most, specific regions of the ER are specialized for Ca2+ storage. Muscle cells, for example, have an abundant specialized smooth ER, called the sarcoplasmic reticulum, which sequesters Ca2+ from the cytosol by means of a Ca2+-ATPase that pumps in Ca2+ into its lumen. The release and reuptake of Ca2+ by the sarcoplasmic reticulum trigger the contraction and relaxation, respectively, of the myofibrils during each round of muscle contraction (discussed in Chapter 16).
We now return to the two major roles of the ER: the synthesis and modification of proteins and the synthesis of lipids.
Rough and Smooth Regions of ER Can Be Separated by Centrifugation
To study the functions and biochemistry of the ER, it is necessary to isolate the ER membrane. This may seem like a hopeless task because the ER is intricately interleaved with other components of the cytosol. Fortunately, when tissues or cells are disrupted by homogenization, the ER breaks into fragments and reseals into many small (~100–200 nm in diameter) closed vesicles called microsomes, which are relatively easy to purify. Microsomes derived from rough ER are studded with ribosomes and are called rough microsomes. The ribosomes are always found on the outside surface, so the interior of the microsome is biochemically equivalent to the lumenal space of the ER (Figure 12-39). Because they can be readily purified in functional form, rough microsomes are especially useful for studying the many processes performed by the rough ER. To the biochemist they represent small authentic versions of the rough ER, still capable of protein synthesis, protein glycosylation, Ca2+ uptake, and lipid synthesis.

Figure 12-39
The isolation of purified rough and smooth microsomes from the ER. (A) When sedimented to equilibrium through a gradient of sucrose, the two types of microsomes separate from each other on the basis of their different densities. (B) A thin section electron (more...)
Many vesicles of a size similar to that of rough microsomes, but lacking attached ribosomes, are also found in these homogenates. Such smooth microsomes are derived in part from smooth portions of the ER and in part from vesiculated fragments of the plasma membrane, Golgi apparatus, endosomes, and mitochondria (the ratio depending on the tissue). Thus, whereas rough microsomes are derived from rough portions of ER, the origins of smooth microsomes cannot be as easily assigned. The microsomes of the liver are an exception. Because of the unusually large quantities of smooth ER in hepatocytes, most of the smooth microsomes in liver homogenates are derived from smooth ER.
The ribosomes attached to rough microsomes make them more dense than smooth microsomes (Figure 12-39B). As a result, the rough and smooth microsomes can be separated from each other by equilibrium centrifugation (see Figure 12-39A). When the separated rough and smooth microsomes of liver are compared with regard to such properties as enzyme activity or polypeptide composition, they are very similar, although not identical: apparently most of the components of the ER membrane can diffuse freely between the rough and smooth regions, as would be expected for a continuous, fluid membrane. The rough microsomes, however, contain more than 20 proteins that are not present in smooth microsomes, showing that some separation mechanism must exist for a subset of ER membrane proteins. Some of the proteins in this subset help to bind ribosomes to the rough ER, while others presumably produce the flattened shape of this part of the ER (see Figure 12-38B). It is not clear whether these membrane proteins are confined to the rough ER by forming large two-dimensional assemblies in the lipid bilayer, or whether they are instead held in place by interactions with a network of structural proteins on one or the other face of the rough ER membrane.
Signal Sequences Were First Discovered in Proteins Imported into the Rough ER
Signal sequences (and the signal sequence strategy of protein sorting) were first discovered in the early 1970s in secreted proteins that are translocated across the ER membrane as a first step toward their eventual discharge from the cell. In the key experiment, the mRNA encoding a secreted protein was translated by ribosomes in vitro. When microsomes were omitted from this cell-free system, the protein synthesized was slightly larger than the normal secreted protein, the extra length being the N-terminal leader peptide. In the presence of microsomes derived from the rough ER, however, a protein of the correct size was produced. These results were explained by the signal hypothesis, which postulated that the leader serves as an ER signal sequence that directs the secreted protein to the ER membrane and is then cleaved off by a signal peptidase in the ER membrane before the polypeptide chain has been completed (Figure 12-40).

Figure 12-40
The signal hypothesis. A simplified view of protein translocation across the ER membrane, as originally proposed. When the ER signal sequence emerges from the ribosome, it directs the ribosome to a translocator on the ER membrane that forms a pore in (more...)
According to the signal hypothesis, the secreted protein should be extruded into the lumen of the microsome during its synthesis in vitro. This can be demonstrated by treatment with a protease: a newly synthesized protein made in the absence of microsomes is degraded when the protease is added to the medium, whereas the same protein made in the presence of microsomes remains intact because it is protected by the microsomal membrane. When proteins without ER signal sequences are similarly synthesized in vitro, they are not imported into microsomes and are therefore degraded by protease treatment.
The signal hypothesis has been thoroughly tested by genetic and biochemical experiments and is found to apply to both plant and animal cells, as well as to protein translocation across the bacterial plasma membrane and, as we have seen, the membranes of mitochondria, chloroplasts, and peroxisomes. N-terminal ER signal sequences guide not only soluble secreted proteins, but also the precursors of all other proteins made by ribosomes bound to the rough ER membrane, including membrane proteins. The signaling function of these peptides has been demonstrated directly by using recombinant DNA techniques to attach ER signal sequences to proteins that do not normally have them; the resulting fusion proteins are directed to the ER.
Cell-free systems in which proteins are imported into microsomes have provided powerful assay procedures for identifying, purifying, and studying the various components of the molecular machinery responsible for the ER import process.
A Signal-Recognition Particle (SRP) Directs ER Signal Sequences to a Specific Receptor in the Rough ER Membrane
The ER signal sequence is guided to the ER membrane by at least two components: a signal-recognition particle (SRP), which cycles between the ER membrane and the cytosol and binds to the signal sequence, and an SRP receptor in the ER membrane. The SRP is a complex particle consisting of six different polypeptide chains bound to a single small RNA molecule (Figure 12-41A). Homologs of the SRP and its receptor are found in all organisms that have been studied, indicating that this protein-targeting mechanism arose early in evolution and has been conserved.

Figure 12-41
The signal-recognition particle (SRP). (A) A mammalian SRP is an elongated complex containing six protein subunits and one RNA molecule (SRP RNA). One end of the SRP binds to an ER signal sequence on a growing polypeptide chain, while the other end binds (more...)
ER signal sequences vary greatly in amino acid sequence, but each has eight or more nonpolar amino acids at its center (see Table 12-3, p. 667). How can the SRP bind specifically to so many different sequences? The answer has come from the crystal structure of the SRP protein, which shows that the signal-sequence-binding site is a large hydrophobic pocket lined by methionines (Figure 12-41B). Because methionines have an unbranched, flexible side chains, the pocket is sufficiently plastic to accommodate hydrophobic signal sequences of different sequences and shapes.
The SRP binds to the ER signal sequence as soon as the peptide has emerged from the ribosome. This causes a pause in protein synthesis, the pause presumably gives the ribosome enough time to bind to the ER membrane before the synthesis of the polypeptide chain is completed, thereby ensuring that the protein is not released into the cytosol. This safety device may be especially important for secreted and lysosomal hydrolases that could wreak havoc in the cytosol; however, cells that secrete large amounts of hydrolases take the added precaution of having high concentrations of hydrolase inhibitors in their cytosol.
Once formed, the SRP-ribosome complex binds to the SRP receptor, which is an integral membrane protein exposed only on the cytosolic surface of the rough ER membrane. This interaction brings the SRP-ribosome complex to a protein translocator. The SRP and SRP receptor are then released, and the growing polypeptide chain is transferred across the membrane (Figure 12-42).

Figure 12-42
How ER signal sequences and SRP direct ribosomes to the ER membrane. The SRP and its receptor are thought to act in concert. The SRP binds to both the exposed ER signal sequence and the ribosome, thereby inducing a pause in translation. The SRP receptor (more...)
The Polypeptide Chain Passes Through an Aqueous Pore in the Translocator
It has long been debated whether polypeptide chains are transferred across the ER membrane in direct contact with the lipid bilayer or through a pore in a protein translocator. The debate ended with the purification of the protein translocator, which was shown to form a water-filled pore in the membrane through which the polypeptide chain traverses the membrane. The translocator, called the Sec61 complex, consists of three or four protein complexes, each composed of three transmembrane proteins, that assemble into a donutlike structure.
When a ribosome binds, the central hole in the translocator lines up with a tunnel in the large ribosomal subunit through which the growing polypeptide chain exits from the ribosome (Figure 12-43). The bound ribosome forms a tight seal with the translocator, such that the space inside the ribosome is continuous with the lumen of the ER and no molecules can escape from the ER (Figure 12-44). The pore in the translocator cannot be open permanently, however; if it were, Ca2+ would leak out of the ER when the ribosome detaches. It is thought that a lumenal ER protein serves as a plug or that the translocator itself can rearrange to close the pore when no ribosome is bound. Thus, the pore is a dynamic structure that opens only transiently when a ribosome with a growing polypeptide chain attaches to the ER membrane.

Figure 12-43
A ribosome bound to the Sec61 protein translocator. (A) A reconstruction of the complex from electron microscopic images viewed from the side. (B) A view of the translocator seen from the top (looking down on the membrane). (C) A schematic drawing of (more...)

Figure 12-44
Evidence for a continuous aqueous pore joining the ER lumen and the interior of the ribosome. In this experiment, a fluorescent dye is attached to a portion of the growing polypeptide chain that is still contained within the ribosome. (A) In free ribosomes, (more...)
The signal sequence in the growing polypeptide chain is thought to trigger the opening of the pore: after the signal sequence is released from the SRP and the growing chain has reached a sufficient length, the signal sequence binds to a specific site inside the pore itself, thereby opening the pore. An ER signal sequence is therefore recognized twice: first, by an SRP in the cytosol, and then by a binding site in the ER protein translocator. This may help to ensure that only appropriate proteins enter the lumen of the ER.
Translocation Across the ER Membrane Does Not Always Require Ongoing Polypeptide Chain Elongation
As we have seen, translocation of proteins into mitochondria, chloroplasts, and peroxisomes occurs posttranslationally, after the protein has been made and released into the cytosol, whereas translocation across the ER membrane usually occurs during translation (co-translationally). This explains why ribosomes are bound to the ER but usually not to other organelles.
Some proteins, however, are imported into the ER after their synthesis has been completed, demonstrating that translocation does not always require ongoing translation. Posttranslational protein translocation is especially common across the ER membrane in yeast cells and across the bacterial plasma membrane (which is thought to be evolutionarily related to the ER; see Figure 12-4). To function in posttranslational translocation, the translocator needs accessory proteins that feed the polypeptide chain into the pore and drive translocation (Figure 12-45). In bacteria, a translocation motor protein, the SecA ATPase, attaches to the cytosolic side of the translocator, where it undergoes cyclic conformational changes driven by ATP hydrolysis. Each time an ATP is hydrolyzed, a portion of the SecA protein inserts into the pore of the translocator, pushing a short segment of the passenger protein with it. As a result of this ratchet mechanism, the SecA protein pushes the polypeptide chain of the transported protein across the membrane.
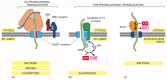
Figure 12-45
Three ways in which protein translocation can be driven through structurally similar translocators. (A) Co-translational translocation. The ribosome is brought to the membrane by the SRP and SRP receptor and forms a tight seal with the Sec61 protein translocator. (more...)
Eucaryotic cells use a different set of accessory proteins that associate with the Sec61 complex. These proteins span the ER membrane and use a small domain on the lumenal side of the ER membrane to deposit an hsp70-like chaperone protein (called BiP, for binding protein) onto the polypeptide chain as it emerges from the pore into the ER lumen. Unidirectional translocation is driven by cycles of BiP binding and release, as described earlier for the mitochondrial hsp70 proteins that pull proteins across mitochondrial membranes.
Proteins that are transported into the ER by a posttranslational mechanism are first released into the cytosol, where they are prevented from folding up by binding to chaperone proteins, as discussed earlier for proteins destined for mitochondria and chloroplasts. In all of these cases where translocation occurs without a ribosome sealing the pore, it remains a mystery how the polypeptide chain can slide through the pore in the translocator without allowing ions and other molecules to pass through.
The ER Signal Sequence Is Removed from Most Soluble Proteins After Translocation
We have seen that in chloroplasts and mitochondria, the signal sequence is cleaved from precursor proteins once it has crossed the membrane. Similarly, N-terminal ER signal sequences are removed by a signal peptidase on the lumenal side of the ER membrane. The signal sequence by itself, however, is not sufficient for signal cleavage by the peptidase; this requires an adjacent cleavage site that is specifically recognized by the peptidase. We shall see below that ER signal sequences that occur within the polypeptide chain—rather than at the N-terminus—do not have these recognition sites and are never cleaved; instead, they can serve to retain transmembrane proteins in the lipid bilayer after the translocation process has been completed.
The N-terminal ER signal sequence of a soluble protein has two signaling functions. It directs the protein to the ER membrane, and it serves as a start-transfer signal (or start-transfer peptide) that opens the pore. Even after it is cleaved off by signal peptidase, the signal sequence is thought to remain bound to the translocator while the rest of the protein is threaded continuously through the membrane as a large loop. Once the C-terminus of the protein has passed through the membrane, the translocated protein is released into the ER lumen (Figure 12-46). The signal sequence is released from the pore and rapidly degraded to amino acids by other proteases in the ER.
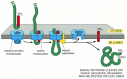
Figure 12-46
A model for how a soluble protein is translocated across the ER membrane. On binding an ER signal sequence (which acts as a start-transfer signal), the translocator opens its pore, allowing the transfer of the polypeptide chain across the lipid bilayer (more...)
While bound in the translocation pore, signal sequences are in contact not only with the Sec61 complex, which forms the walls of the pore, but also with the hydrophobic lipid core of the membrane. This was shown in chemical cross-linking experiments in which signal sequences and the hydrocarbon chains of lipids could be covalently linked together. To release the signal sequence into the membrane, the translocator has to open laterally. The translocator is therefore gated in two directions: it can open to form a pore across the membrane to let the hydrophilic portions of proteins cross the lipid bilayer, and it can open laterally within the membrane to let hydrophobic portions of proteins partition into the bilayer. This lateral gating mechanism is crucial for the insertion of transmembrane proteins into the lipid bilayer, as we discuss next.
In Single-Pass Transmembrane Proteins, a Single Internal ER Signal Sequence Remains in the Lipid Bilayer as a Membrane-spanning α Helix
The translocation process for proteins destined to remain in the membrane is more complex than it is for soluble proteins, as some parts of the polypeptide chain are translocated across the lipid bilayer whereas others are not. Nevertheless, all modes of insertion of membrane proteins can be considered as variants of the sequence of events just described for transferring a soluble protein into the lumen of the ER. We begin by describing the three ways in which single-pass transmembrane proteins (see Figure 10-17) become inserted into the ER.
In the simplest case, an N-terminal signal sequence initiates translocation, just as for a soluble protein, but an additional hydrophobic segment in the polypeptide chain stops the transfer process before the entire polypeptide chain is translocated. This stop-transfer signal anchors the protein in the membrane after the ER signal sequence (the start-transfer signal) has been released from the translocator and has been cleaved off (Figure 12-47). The stop-transfer sequence is transferred into the bilayer by the lateral gating mechanism, and it remains there as a single α-helical membrane-spanning segment, with the N-terminus of the protein on the lumenal side of the membrane and the C-terminus on the cytosolic side.
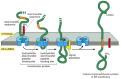
Figure 12-47
How a single-pass transmembrane protein with a cleaved ER signal sequence is integrated into the ER membrane. In this hypothetical protein the co-translational translocation process is initiated by an N-terminal ER signal sequence (red) that functions (more...)
In the other two cases, the signal sequence is internal, rather than at the N-terminal end of the protein. Like the N-terminal ER signal sequences, the internal signal sequence is recognized by an SRP, which brings the ribosome making the protein to the ER membrane and serves as a start-transfer signal that initiates the translocation of the protein. After release from the translocator, the internal start-transfer sequence remains in the lipid bilayer as a single membrane-spanning α helix.
Internal start-transfer sequences, can bind to the translocation apparatus in either of two orientations, and the orientation of the inserted start-transfer sequence, in turn, determines which protein segment (the one preceding or the one following the start-transfer sequence) is moved across the membrane into the ER lumen. In one case, the resulting membrane protein has its C-terminus on the lumenal side (Figure 12-48A), while in the other, it has its N-terminus on the lumenal side (Figure 12-48B). The orientation of the start-transfer sequence depends on the distribution of nearby charged amino acids, as described in the figure legend.
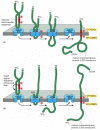
Figure 12-48
Integration of a single-pass membrane protein with an internal signal sequence into the ER membrane. In these hypothetical proteins, an internal ER signal sequence that functions as a start-transfer signal binds to the translocator in such a way that (more...)
Combinations of Start-Transfer and Stop-Transfer Signals Determine the Topology of Multipass Transmembrane Proteins
In multipass transmembrane proteins, the polypeptide chain passes back and forth repeatedly across the lipid bilayer (see Figure 10-17). It is thought that an internal signal sequence serves as a start-transfer signal in these proteins to initiate translocation, which continues until a stop-transfer sequence is reached. In double-pass transmembrane proteins, for example, the polypeptide can then be released into the bilayer (Figure 12-49). In more complex multipass proteins, in which many hydrophobic α helices span the bilayer, a second start-transfer sequence reinitiates translocation further down the polypeptide chain until the next stop-transfer sequence causes polypeptide release, and so on for subsequent start-transfer and stop-transfer sequences (Figure 12-50).
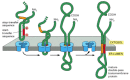
Figure 12-49
Integration of a double-pass membrane protein with an internal signal sequence into the ER membrane. In this hypothetical protein, an internal ER signal sequence acts as a start-transfer signal (as in Figure 12-48) and initiates the transfer of the C-terminal (more...)

Figure 12-50
The insertion of the multipass membrane protein rhodopsin into the ER membrane. Rhodopsin is the light-sensitive protein in rod photoreceptor cells in the mammalian retina (discussed in Chapter 15). (A) A hydrophobicity plot identifies seven short hydrophobic (more...)
Whether a given hydrophobic signal sequence functions as a start-transfer or stop-transfer sequence must depend on its location in a polypeptide chain, since its function can be switched by changing its location in the protein using recombinant DNA techniques. Thus, the distinction between start-transfer and stop-transfer sequences results mostly from their relative order in the growing polypeptide chain. It seems that the SRP begins scanning an unfolded polypeptide chain for hydrophobic segments at its N-terminus and proceeds toward the C-terminus, in the direction that the protein is synthesized. By recognizing the first appropriate hydrophobic segment to emerge from the ribosome, the SRP sets the “reading frame”: if translocation is initiated, the next appropriate hydrophobic segment is recognized as a stop-transfer sequence, causing the region of the polypeptide chain in between to be threaded across the membrane. A similar scanning process continues until all of the hydrophobic regions in the protein have been inserted into the membrane.
Because membrane proteins are always inserted from the cytosolic side of the ER in this programmed manner, all copies of the same polypeptide chain will have the same orientation in the lipid bilayer. This generates an asymmetrical ER membrane in which the protein domains exposed on one side are different from those domains exposed on the other. This asymmetry is maintained during the many membrane budding and fusion events that transport the proteins made in the ER to other cell membranes (discussed in Chapter 13). Thus, the way in which a newly synthesized protein is inserted into the ER membrane determines the orientation of the protein in all of the other membranes as well.
When proteins are dissociated from a membrane and are then reconstituted into artificial lipid vesicles, a random mixture of right-side-out and inside-out protein orientations usually results. Thus, the protein asymmetry observed in cell membranes seems not to be an inherent property of the protein, but instead results solely from the process by which proteins are inserted into the ER membrane from the cytosol.
Translocated Polypeptide Chains Fold and Assemble in the Lumen of the Rough ER
Many of the proteins in the lumen of the ER are in transit, en route to other destinations; others, however, are normally resident there and are present at high concentrations. These ER resident proteins contain an ER retention signal of four amino acids at their C terminus that is responsible for retaining the protein in the ER (see Table 12-3; discussed in Chapter 13). Some of these proteins function as catalysts that help the many proteins that are translocated into the ER to fold and assemble correctly.
One important ER resident protein is protein disulfide isomerase (PDI), which catalyzes the oxidation of free sulfhydryl (SH) groups on cysteines to form disulfide (S-S) bonds. Almost all cysteines in protein domains exposed to either the extracellular space or the lumen of organelles in the secretory and endocytic pathways are disulfide-bonded; disulfide bonds do not form, however, in domains exposed to the cytosol because of the reducing environment there.
Another ER resident protein is the chaperone protein BiP. We have already discussed how BiP works to pull proteins posttranslationally into the ER through the ER translocator. Like other chaperones, BiP recognizes incorrectly folded proteins, as well as protein subunits that have not yet assembled into their final oligomeric complexes. To do so, it binds to exposed amino acid sequences that would normally be buried in the interior of correctly folded or assembled polypeptide chains. An example of a BiP-binding site is a stretch of alternating hydrophobic and hydrophilic amino acids that would normally be buried in a β sheet. The bound BiP both prevents the protein from aggregating and helps to keep it in the ER (and thus out of the Golgi apparatus and later parts of the secretory pathway). Like the hsp70 family of proteins, which bind unfolded proteins in the cytosol and facilitate their import into mitochondria and chloroplasts, BiP hydrolyzes ATP to provide the energy for its roles in protein folding and posttranslational import into the ER.
Most Proteins Synthesized in the Rough ER Are Glycosylated by the Addition of a Common N-linked Oligosaccharide
The covalent addition of sugars to proteins is one of the major biosynthetic functions of the ER. Most of the soluble and membrane-bound proteins that are made in the ER—including those destined for transport to the Golgi apparatus, lysosomes, plasma membrane, or extracellular space—are glycoproteins. In contrast, very few proteins in the cytosol are glycosylated, and those that are carry a much simpler sugar modification, in which a single N-acetylglucosamine group is added to a serine or threonine residue of the protein.
An important advance in understanding the process of protein glycosylation was the discovery that a preformed precursor oligosaccharide (composed of N-acetylglucosamine, mannose, and glucose and containing a total of 14 sugars) is transferred en bloc to proteins in the ER. Because this oligosaccharide is transferred to the side-chain NH2 group of an asparagine amino acid in the protein, it is said to be N-linked or asparagine-linked (Figure 12-51). The transfer is catalyzed by a membrane-bound enzyme, an oligosaccharyl transferase, which has its active site exposed on the lumenal side of the ER membrane; this explains why cytosolic proteins are not glycosylated in this way. The precursor oligosaccharide is held in the ER membrane by a special lipid molecule called dolichol, and it is transferred to the target asparagine in a single enzymatic step immediately after that amino acid has emerged into the ER lumen during protein translocation (Figure 12-52). Since most proteins are co-translationally imported into the ER, N-linked oligosaccharides are almost always added during protein synthesis.

Figure 12-51
The asparagine-linked (N-linked) precursor oligosaccharide that is added to most proteins in the rough ER membrane. The five sugars in the gray box form the “core region” of this oligosaccharide. For many glycoproteins, only the core (more...)
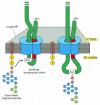
Figure 12-52
Protein glycosylation in the rough ER. Almost as soon as a polypeptide chain enters the ER lumen, it is glycosylated on target asparagine amino acids. The precursor oligosaccharide shown in Figure 12-51 is transferred to the asparagine as an intact unit (more...)
The precursor oligosaccharide is linked to the dolichol lipid by a high-energy pyrophosphate bond, which provides the activation energy that drives the glycosylation reaction illustrated in Figure 12-52. The entire precursor oligosaccharide is built up sugar by sugar on this membrane-bound lipid molecule before its transfer to a protein. The sugars are first activated in the cytosol by the formation of nucleotide-sugar intermediates, which then donate their sugar (directly or indirectly) to the lipid in an orderly sequence. Partway through this process, the lipid-linked oligosaccharide is flipped from the cytosolic to the lumenal side of the ER membrane (Figure 12-53).

Figure 12-53
Synthesis of the lipid-linked precursor oligosaccharide in the rough ER membrane. The oligosaccharide is assembled sugar by sugar onto the carrier lipid dolichol (a polyisoprenoid; see Panel 2-5, pp. 118–119). Dolichol is long and very hydrophobic: (more...)
All of the diversity of the N-linked oligosaccharide structures on mature glycoproteins results from the later modification of the original precursor oligosaccharide. While still in the ER, three glucoses (see Figure 12-51) and one mannose are quickly removed from the oligosaccharides of most glycoproteins. We shall return to the importance of glucose trimming shortly. This oligosaccharide “trimming” or “processing” continues in the Golgi apparatus and is discussed in Chapter 13.
The N-linked oligosaccharides are by far the most common oligosaccharides found on glycoproteins. Less frequently, oligosaccharides are linked to the hydroxyl group on the side chain of a serine, threonine, or hydroxylysine amino acid. These O-linked oligosaccharides are formed in the Golgi apparatus by pathways that are not yet fully understood.
Oligosaccharides Are Used as Tags to Mark the State of Protein Folding
It has long been debated why glycosylation is such a common modification of proteins that enter the ER. One particularly puzzling observation has been that some proteins require N-linked glycosylation for proper folding in the ER, yet the precise location of the oligosaccharides attached to the protein's surface does not seem to matter. A clue to the role of glycosylation in protein folding came from studies of two ER chaperone proteins that are called calnexin and calreticulin because they require Ca2+ for their activities. These chaperones are lectins that bind to oligosaccharides on incompletely folded proteins and retain them in the ER. Like other chaperones, they prevent incompletely folded proteins from undergoing irreversible aggregation. Both calnexin and calreticulin also promote the association of incompletely folded protein with another ER chaperone, which binds to cysteines that have not yet formed disulfide bonds.
Calnexin and calreticulin recognize N-linked oligosaccharides that contain a single terminal glucose, and therefore bind proteins only after two of the three glucoses that are initially attached have been removed by ER glucosidases. When the third glucose is removed, the protein dissociates from its chaperone and can leave the ER.
How, then, do calnexin and calreticulin distinguish folded from incompletely folded proteins? The answer lies in yet another ER enzyme, a glucosyl transferase that keeps adding a glucose to those oligosaccharides that have lost their last glucose. It adds the glucose, however, only to oligosaccharides that are attached to unfolded proteins. Thus, an unfolded protein undergoes continuous cycles of glucose trimming (by glucosidase) and addition (by glycosyl transferase), and maintains an affinity for calnexin and calreticulin until it has achieved its fully folded state (Figure 12-54).

Figure 12-54
The role of N-linked glycosylation in ER protein folding. The ER-membrane-bound chaperone protein calnexin binds to incompletely folded proteins containing one terminal glucose on N-linked oligosaccharides, trapping the protein in the ER. Removal of the (more...)
Improperly Folded Proteins Are Exported from the ER and Degraded in the Cytosol
Despite all the help from chaperones, many protein molecules (more than 80% for some proteins) translocated into the ER fail to achieve their properly folded or oligomeric state. Such proteins are exported from the ER back into the cytosol, where they are degraded. The retrotranslocation, also called dislocation, occurs via the same translocator (the Sec61 complex) through which the proteins entered the ER in the first place, although additional proteins help the translocator to function in reverse. It is not known how such misfolded proteins, which no longer have their ER signal sequences, are recognized or transferred.
Once the misfolded protein has reached the cytosol, its oligosaccharides are removed. Deglycosylation is catalyzed by an N-glycanase, which removes the oligosaccharide chains by cleaving the amide bond between the carbonyl group and the amino group of the original asparagine to which the oligosaccharide was attached. The deglycosylated polypeptide is rapidly ubiquitylated by ER-bound ubiquitin-conjugating enzymes and is then fed into proteasomes (discussed in Chapter 6), where it is degraded (Figure 12-55).

Figure 12-55
The export and degradation of misfolded ER proteins. Misfolded soluble proteins in the ER lumen are translocated back into the cytosol, where they are deglycosylated, ubiquitylated, and degraded in proteasomes. Misfolded membrane proteins follow a similar (more...)
Misfolded Proteins in the ER Activate an Unfolded Protein Response
Cells carefully monitor the amount of misfolded proteins they contain in various compartments. An accumulation of misfolded proteins in the cytosol, for example, triggers a heat-shock response (discussed in Chapter 6), which stimulates the transcription of genes encoding cytosolic chaperones that help to refold the proteins. Similarly, an accumulation of misfolded proteins in the ER triggers an unfolded protein response, which includes an increased transcription of genes encoding ER chaperones and enzymes involved in ER protein degradation.
How do misfolded proteins in the cytosol or ER signal to the nucleus? The pathway from the ER to the nucleus is especially well understood in yeast cells, and it is remarkable. A transmembrane protein kinase in the ER is activated by misfolded proteins, which cause its oligomerization and autophosphorylation. (Extracellular growth factors activate their receptors in the plasma membrane in a similar way, as discussed in Chapter 15). Oligomerization of the ER kinase leads to the activation of an endoribonuclease domain contained on the same molecule. This nuclease cleaves a specific, cytosolic RNA molecule at two positions, excising an intron. The separated exons are then joined by an RNA ligase, generating a spliced mRNA, which is translated on ribosomes to produce a gene regulatory protein. The protein migrates to the nucleus and activates the transcription of the genes encoding the proteins that mediate the unfolded protein response (Figure 12-56).

Figure 12-56
The unfolded protein response in yeast. By this novel intracellular signaling pathway, the accumulation of misfolded proteins in the ER lumen signals to the nucleus to activate the transcription of genes that encode proteins that help the cell to cope (more...)
Some Membrane Proteins Acquire a Covalently Attached Glycosylphosphatidylinositol (GPI) Anchor
As discussed in Chapter 10, several cytosolic enzymes catalyze the covalent addition of a single fatty acid chain or prenyl group to selected proteins. The attached lipids help to direct these proteins to cell membranes. A related process is catalyzed by ER enzymes, which covalently attach a glycosylphosphatidyl-inositol (GPI) anchor to the C terminus of some membrane proteins destined for the plasma membrane. This linkage forms in the lumen of the ER, where, at the same time, the transmembrane segment of the protein is cleaved off (Figure 12-57). A large number of plasma membrane proteins are modified in this way. Since they are attached to the exterior of the plasma membrane only by their GPI anchors, they can in principle be released from cells in soluble form in response to signals that activate a specific phospholipase in the plasma membrane. Trypanosome parasites, for example, use this mechanism to shed their coat of GPI-anchored surface proteins if attacked by the immune system. GPI anchors are also used to direct plasma membrane proteins into lipid rafts and thus segregate the proteins from other membrane proteins, as we discuss in Chapter 13.

Figure 12-57
The attachment of a GPI anchor to a protein in the ER. Immediately after the completion of protein synthesis, the precursor protein remains anchored in the ER membrane by a hydrophobic C-terminal sequence of 15–20 amino acids; the rest of the (more...)
Most Membrane Lipid Bilayers Are Assembled in the ER
The ER membrane synthesizes nearly all of the major classes of lipids, including both phospholipids and cholesterol, required for the production of new cell membranes. The major phospholipid made is phosphatidylcholine (also called lecithin), which can be formed in three steps from choline, two fatty acids, and glycerol phosphate (Figure 12-58). Each step is catalyzed by enzymes in the ER membrane that have their active sites facing the cytosol, where all of the required metabolites are found. Thus, phospholipid synthesis occurs exclusively in the cytosolic leaflet of the ER membrane. In the first step, acyl transferases successively add two fatty acids to glycerol phosphate to produce phosphatidic acid, a compound sufficiently water-insoluble to remain in the lipid bilayer after it has been synthesized. It is this step that enlarges the lipid bilayer. The later steps determine the head group of a newly formed lipid molecule, and therefore the chemical nature of the bilayer, but they do not result in net membrane growth. The two other major membrane phospholipids—phosphatidyl-ethanolamine and phosphatidylserine—as well as the minor phospholipid phosphatidylinositol (PI), are all synthesized in this way.

Figure 12-58
The synthesis of phosphatidylcholine. This phospholipid is synthesized from fatty acyl-coenzyme A (fatty acyl CoA), glycerol 3-phosphate, and cytidine-bisphosphocholine (CDP-choline).
Because phospholipid synthesis takes place in the cytosolic half of the ER bilayer, there needs to be a mechanism that transfers some of the newly formed phospholipid molecules to the lumenal leaflet of the bilayer. In synthetic lipid bilayers, lipids do not “flip-flop” in this way. In the ER, however, phospholipids equilibrate across the membrane within minutes, which is almost 100,000 times faster than can be accounted for by spontaneous “flip-flop.” This rapid trans-bilayer movement is thought to be mediated by a phospholipid translocator called a scramblase that equilibrates phospholipids between the two leaflets of the lipid bilayer (Figure 12-59). Thus, the different types of phospholipids are thought to be equally distributed between the two leaflets of the ER membrane. The plasma membrane contains, in addition to the scramblase, a different type of phospholipid translocator that belongs to the family of ABC transporters (discussed in Chapter 11). These flippases specifically remove phospholipids containing free amino groups (phosphatidylserine and phosphatidylethanolamine) from the extracellular leaflet and use the energy of ATP hydrolysis to flip them directionally into the leaflet facing the cytosol. The plasma membrane therefore has a highly asymmetric phospholipid composition, which is actively maintained by the flippases (see Figure 10-14).

Figure 12-59
The role of phospholipid translocators in lipid bilayer synthesis. (A) Because new lipid molecules are added only to the cytosolic half of the bilayer and lipid molecules do not flip spontaneously from one monolayer to the other, a membrane-bound phospholipid (more...)
The ER also produces cholesterol and ceramide. Ceramide is made by condensing the amino acid serine with a fatty acid to form the amino alcohol sphingosine; a second fatty acid is then added to form ceramide. The ceramide is exported to the Golgi apparatus, where it serves as a precursor for the synthesis of two types of lipids: oligosaccharide chains are added to form glycosphingo-lipids (glycolipids), and phosphocholine head groups are transferred from phosphatidylcholine to other ceramide molecules to form sphingomyelin. Thus, both glycolipids and sphingomyelin are produced relatively late in the process of membrane synthesis. Because they are produced by enzymes exposed to the Golgi lumen and are not substrates for lipid translocators, they are found exclusively in the noncytosolic leaflet of the lipid bilayers that contain them.
Phospholipid Exchange Proteins Help to Transport Phospholipids from the ER to Mitochondria and Peroxisomes
As discussed in Chapter 13, the plasma membrane and the membranes of the Golgi apparatus, lysosomes, and endosomes all form part of a membrane system that communicates with the ER by means of transport vesicles that transfer both proteins and lipids. Mitochondria, plastids, and possibly peroxisomes, however, do not belong to this system, and they therefore require different mechanisms for the import of proteins and lipids for growth. We have already seen that most of the proteins in these organelles are imported from the cytosol. Although mitochondria modify some of the lipids they import, they do not synthesize lipids from scratch; instead, their lipids have to be imported from the ER, either directly, or indirectly by way of other cell membranes. In either case, special mechanisms are required for the transfer.
Water-soluble carrier proteins called phospholipid exchange proteins (or phospholipid transfer proteins) transfer individual phospholipid molecules between membranes. Each exchange protein recognizes only specific types of phospholipids. It functions by “extracting” a molecule of the appropriate phospholipid from a membrane and diffusing away with the lipid buried within its lipid-binding site. When it encounters another membrane, the exchange protein tends to discharge the bound phospholipid molecule into the new lipid bilayer (Figure 12-60). It has been proposed that phosphatidylserine is imported into mitochondria in this way, where it is then decarboxylated to yield phosphatidylethanolamine. Phosphatidylcholine, by contrast, is imported intact.
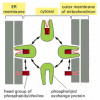
Figure 12-60
Phospholipid exchange proteins. Because phospholipids are insoluble in water, their passage between membranes requires carrier proteins. Phospholipid exchange proteins are water-soluble proteins that carry a single molecule of phospholipid at a time; (more...)
Exchange proteins act to distribute phospholipids at random between all membranes present. In principle, such a random exchange process can result in a net transport of lipids from a lipid-rich to a lipid-poor membrane, allowing phosphatidylcholine and phosphatidylserine molecules, for example, to be transferred from the ER, where they are synthesized, to a mitochondrial or peroxisomal membrane. It might be that mitochondria and peroxisomes are the only “lipid-poor” organelles in the cytosol and that such an exchange process is sufficient. In electron micrographs, mitochondria are often seen in close juxtaposition to ER membranes, and there may be specific mechanisms of lipid transfer that operate at such regions of proximity.
Summary
The extensive ER network serves as a factory for the production of almost all of the cell's lipids. In addition, a major portion of the cell's protein synthesis occurs on the cytosolic surface of the ER: all proteins destined for secretion and all proteins destined for the ER itself, the Golgi apparatus, the lysosomes, the endosomes, and the plasma membrane are first imported into the ER from the cytosol. In the ER lumen, the proteins fold and oligomerize, disulfide bonds are formed, and N-linked oligosaccharides are added. N-linked glycosylation is used to indicate the extent of protein folding, so that proteins leave the ER only when they are properly folded. Proteins that do not fold or oligomerize correctly are translocated back into the cytosol, where they are deglycosylated, ubiquitylated, and degraded in proteasomes. If misfolded proteins accumulate excessively in the ER, they trigger an unfolded protein response, which activates appropriate genes in the nucleus to help the ER to cope.
Only proteins that carry a special ER signal sequence are imported into the ER. The signal sequence is recognized by a signal recognition particle (SRP), which binds both the growing polypeptide chain and a ribosome and directs them to a receptor protein on the cytosolic surface of the rough ER membrane. This binding to the ER membrane initiates the translocation process by threading a loop of polypeptide chain across the ER membrane through the hydrophilic pore in a transmembrane protein translocator.
Soluble proteins—destined for the ER lumen, for secretion, or for transfer to the lumen of other organelles—pass completely into the ER lumen. Transmembrane proteins destined for the ER or for other cell membranes are translocated partway across the ER membrane and remain anchored there by one or more membrane-spanning α-helical regions in their polypeptide chains. These hydrophobic portions of the protein can act either as start-transfer or stop-transfer signals during the translocation process. When a polypeptide contains multiple, alternating start-transfer and stop-transfer signals, it will pass back and forth across the bilayer multiple times as a multipass transmembrane protein.
The asymmetry of protein insertion and glycosylation in the ER establishes the sidedness of the membranes of all of the other organelles that the ER supplies with membrane proteins.
- Membrane-bound Ribosomes Define the Rough ER
- Smooth ER Is Abundant in Some Specialized Cells
- Rough and Smooth Regions of ER Can Be Separated by Centrifugation
- Signal Sequences Were First Discovered in Proteins Imported into the Rough ER
- A Signal-Recognition Particle (SRP) Directs ER Signal Sequences to a Specific Receptor in the Rough ER Membrane
- The Polypeptide Chain Passes Through an Aqueous Pore in the Translocator
- Translocation Across the ER Membrane Does Not Always Require Ongoing Polypeptide Chain Elongation
- The ER Signal Sequence Is Removed from Most Soluble Proteins After Translocation
- In Single-Pass Transmembrane Proteins, a Single Internal ER Signal Sequence Remains in the Lipid Bilayer as a Membrane-spanning α Helix
- Combinations of Start-Transfer and Stop-Transfer Signals Determine the Topology of Multipass Transmembrane Proteins
- Translocated Polypeptide Chains Fold and Assemble in the Lumen of the Rough ER
- Most Proteins Synthesized in the Rough ER Are Glycosylated by the Addition of a Common N-linked Oligosaccharide
- Oligosaccharides Are Used as Tags to Mark the State of Protein Folding
- Improperly Folded Proteins Are Exported from the ER and Degraded in the Cytosol
- Misfolded Proteins in the ER Activate an Unfolded Protein Response
- Some Membrane Proteins Acquire a Covalently Attached Glycosylphosphatidylinositol (GPI) Anchor
- Most Membrane Lipid Bilayers Are Assembled in the ER
- Phospholipid Exchange Proteins Help to Transport Phospholipids from the ER to Mitochondria and Peroxisomes
- Summary
- The Endoplasmic Reticulum - Molecular Biology of the CellThe Endoplasmic Reticulum - Molecular Biology of the Cell
Your browsing activity is empty.
Activity recording is turned off.
See more...