All Jasper's Basic Mechanisms of the Epilepsies content, except where otherwise noted, is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported license, which permits copying, distribution and transmission of the work, provided the original work is properly cited, not used for commercial purposes, nor is altered or transformed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Noebels JL, Avoli M, Rogawski MA, et al., editors. Jasper's Basic Mechanisms of the Epilepsies [Internet]. 4th edition. Bethesda (MD): National Center for Biotechnology Information (US); 2012.
This title is an author manuscript version first made accessible on the NCBI Bookshelf website July 2, 2012.
Infantile epileptic encephalopathies, such as Dravet syndrome, Ohtahara syndrome, West syndrome, Lennox-Gastaut syndrome, myoclonic-astatic epilepsy, and Landau-Kleffner syndrome, are devastating epilepsies. Cases are often sporadic or patients have only a limited family history of epilepsy. Although a complex inheritance has long been suspected in epilepsy, recent data indicate that many sporadic rare epileptic disorders, such as Dravet syndrome, CDKL5/STK9 Rett-like epileptic encephalopathy, ARX-related epilepsies, SRPX2-related rolandic epilepsy associated with oral and speech dyspraxia and mental retardation, and STXBP1-related West/Ohtahara syndromes, are due to a mutation in a unique gene. Dravet syndrome, for example, is mainly due to de novo mutations in SCN1A, the gene encoding the voltage-gated neuronal sodium channel alpha 1 subunit, which explains why most patients are isolated cases. All types of mutations are observed: missense mutations, premature termination codon and intragenic rearrangements. This large mutation spectrum contrasts with that of generalized epilepsy with febrile seizures plus (GEFS+), an autosomal dominant condition also characterized by febrile and afebrile seizures but with a usually benign outcome, in which only missense mutations are found. Recently, mutations in PCDH19, encoding protocadherin 19 on chromosome X, were identified in females with an EFMR or Dravet-like phenotype. Heterozygous females are affected while hemizygous males are spared, this unusual inheritance probably being due to a mechanism called cellular interference. The genetic data accumulated for Dravet syndrome and other related disorders have to be kept in mind when studying epileptic encephalopathies.
Epileptic encephalopathies (EEs) are conditions in which cognitive, sensory, and/or motor function deterioration results mainly from epileptic activity.1 This epileptic activity, which can be frequent and severe as in Dravet syndrome or continuous, subcontinuous or abundant in the “interictal” period as in Landau Kleffner or Lennox-Gastaut syndrome, is believed to have a deleterious impact on the developing and mature brain, interfering with cognitive functions at different times of life. 2
EEs have numerous etiologies, which differ from one to another. 3 For example, various causes were identified in West syndrome, including damage and malformations of the brain and, more rarely, an inborn error of metabolism. However, at least one third are cryptogenic, without any identifiable underlying cause. 2 When the more frequent etiologies have been excluded by extensive biological and neuroimaging examinations, the possibility of a genetic cause must be addressed. In contrast, Dravet syndrome is the archetype of the genetically determined epileptic encephalopathy, with a mutation identified in 60–80% of patients. In this context, genetic testing has to be provided in order to confirm the clinical diagnosis.
Patients with epileptic encephalopathy are mainly sporadic cases, making the differentiation between an acquired or genetic cause a challenge. In rare cases, however, the recurrence of the same phenotype in a family strongly supports the involvement of a mutated gene. In addition, because of the high phenotypic variability of genetic diseases, EE can occur in a familial context of epilepsy. Indeed, 10% of patients with Dravet syndrome have a familial history of epilepsy or febrile convulsions.
In this chapter, we will focus on genes responsible for infantile epileptic encephalopathies as the main component of the clinical picture and not on those included in complex phenotypes encountered in metabolic diseases or in contiguity syndromes due to large chromosomal abnormalities. Indeed, the association of EE with a dysmorphy or extra-neurological symptoms should lead to specific metabolic or cytogenetic investigations being performed. In the first part of this chapter, we will review the genes involved in early infantile epileptic encephalopathies (EIEEs). Although the mutations in these genes are rare, their screening has to be integrated in a rational diagnostic strategy. It is particularly important to rule out genetic etiologies since there is a risk of the disease recurring in the family. In this review, we will focus on Dravet syndrome as an example of a genetically determined infantile EE to emphasize the difficulties and pitfalls that might be encountered in such diseases.
THE MONOGENIC FORMS OF EARLY INFANTILE EPILEPTIC ENCEPHALOPATHIES
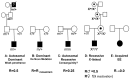
Geneticists have long known that a fraction of sporadic cases of patients with developmental defects could be due to a mutation in a single gene (monogenic forms). All modes of inheritance can be hypothesized, each of them being associated with particular features (Figure 1). Three of them have already been reported in EIEEs: autosomal dominant (mainly de novo mutations), autosomal recessive and X-linked. A genetic classification exists for genetically determined EIEE (OMIM: Online Mendelian Inheritance in Man http://www.ncbi.nlm.nih.gov/omim): EIEE1 (MIM# 3008350) is part of the phenotypic spectrum of disorders caused by mutation in the ARX gene comprising a nearly continuous series of developmental disorders; EIEE2 (MIM# 300672), an X-linked disorder caused by mutation in the CDKL5/STK9 gene (MIM# 300203); EIEE3 (MIM# 609304), caused by mutation in the CG1/SLC25A22 gene (MIM# 609302); EIEE4 (MIM# 612164), caused by mutation in the STXBP1 gene (MIM# 602926); and EIEE5 (MIM# 613477), caused by mutation in the SPTAN1 gene (MIM# 182810). This genetic classification differs from the clinical classification of epileptic encephalopathies based on type of seizures, age at onset, evolution and prognosis (Table 1). 3 The articulation of clinical and genetic classifications will be a challenge in the coming years. The main problem is that the number of mutations identified in each gene is relatively small and is usually associated with a large phenotypic spectrum, making positive phenotype-genotype correlations difficult.
Table 1
Main etiologies of epileptic encephalopathies.
Dominant mutations responsible for EIEE mostly occur de novo
The dominant mutations of STXBP1 responsible for EIEE4
Dominant mutations of STXPB1 (also called MUNC18-1) gene, which is located on chromosome 9q34.11, can cause sporadic EE. In autosomal dominant diseases, a weak penetrance (proportion of affected individuals among mutation carriers) can explain why parents are not both affected, one of them being an asymptomatic carrier. However, for deleterious phenotypes like EEs, most sporadic cases are due to de novo mutations. In this case, mutations that are eliminated from the population by their negative selective effect on patients’ reproduction are counter-balanced by the occurrence of neomutations. The mutation generally occurs in a gamete that will be involved later on in fecundation, the corresponding zygote being affected since the mutation is dominant. This is often the case for EE associated with mutations in STXBP1. STXBP1 gene contains 20 exons and encodes the syntaxin-binding protein 1. Forming a complex with the SNARE protein syntaxin1, the STXBP1 protein might mediate synaptic vesicle fusion. 4 In accordance with this hypothesis, deletion of Munc18-1 (paralog of STXBP1) in mice renders the brain synaptically silent, identifying Munc18-1 (STXBP1) as the currently most upstream essential protein in neurotransmitter release. 5 Saitsu and colleagues (2008) first described mutations in this gene in 5 unrelated cases among 13 patients with EIEE with suppression-burst (Ohtahara syndrome) for whom the main causes of EEIE were excluded. Missense, premature termination codon (non-sense mutations) and deletions were reported, leading to the conclusion that the disease is likely due to a haploinsufficiency (loss of function of the mutated copy of the gene) of STXBP1. 6, 7 EEIE4 is most often associated with neurological signs such as mental retardation, which can be profound, and spastic paraplegia. The rarity of reports on mutations in this gene has to be underlined and raises questions as to whether its systematic screening is indicated for patients with mental retardation and EIEE with suppression-burst.
SPTAN1, a newly identified gene responsible for sporadic EIEE5
Four patients presenting with early onset epileptic encephalopathy with poor attention, severe hypomyelination and reduction in cerebral white matter (EIEE5; 613477), were reported in 2008. On EEG, one patient presented with suppression-burst and the other three had hypsarrhythmia. The authors stated that this phenotype could be considered as a new clinical condition associated with early onset West syndrome (WS). 8, 9 A de novo 9q33.3-q34.11 microdeletion encompassing the STXBP1 gene has been detected in one of these patients. Since no mutations of STXBP1 were found in two of the remaining three subjects, the authors suspected that another gene within the deletion might contribute to this severe phenotype. SPTAN1 encoding alpha-II spectrin, which is involved in cell proliferation via arrest at cell cycle phase G1 and in myelination in zebrafish, 10 was localized in the deletion. In two subjects, an in-frame 3 bp deletion and a 6 bp duplication in SPTAN1, likely resulting in the synthesis of an abnormal protein, were found at the initial nucleation site of the alpha/beta spectrin heterodimer. 9 SPTAN1 was further screened in six unrelated individuals with WS and hypomyelination, but no mutations were found. In vitro functional expression studies suggested a dominant-negative effect of the two identified mutations on spectrin heterodimer stability, as well as perturbation of the axon initial segment.
CG1/SLC25A22, a gene responsible for rare autosomal recessive (AR) EIEE with suppression-burst pattern
In Europe, the vast majority of families with an AR disease have a single affected child because of the small number of siblings. However, for rare diseases, consanguinity has to be searched for since it increases the probability of encountering two mutated copies of a gene in the same individual. This was the case for recessive mutations of GC1 (Glutamate carrier 1; also called SLC25A22 for solute carrier family 25) identified in patients with EIEE3 (EIEE3; OMIM 6093304). This gene (OMIM; 609304) was first mapped on chromosome 11p15.5 in a consanguineous Arab Muslim family from Jerusalem. Patients presented with neonatal intractable seizures associated with a suppression-burst EEG pattern and hypotonia. Brain atrophy was later diagnosed at CT scan (at about 3 years of age). 11 The missense mutation p.Pro206Leu was subsequently identified at the homozygous state in the affected members of this family in the CG1/SLC25A22 gene, which was located within the candidate genomic region. A second mutation, p.Gly236Trp (G236W), which confirmed the responsibility of the CG1 gene in this EE, was more recently identified in an Algerian boy also born from related parents with a severe encephalopathy and without any psychomotor acquisition. 12 GC1/SLC25A22 encodes one of the two mitochondrial glutamate/H+ symporters, the other being SLC25A18. 13 In vitro functional expression assays of G236W-mutated GC1 cDNA in E. coli showed that the mutant protein was correctly inserted into the liposomal membrane but had no functional transport activity, confirming the deleterious consequences of the mutation on the protein function. 12
X-linked epileptic encephalopathies
The X-linked heredity is characterized by a difference of phenotype related to sex. The recessive X-linked mode of inheritance is most often characterized by a familial context, with affected males being born to asymptomatic female carriers. The transmission of the mutation by females over several generations leads to isolated affected males. For severe diseases, sporadic cases can also be encountered by de novo mutation, such as in Duchenne muscular dystrophy in which one third of affected males are sporadic cases due to de novo mutations. In dominant X-linked inheritance, heterozygous females can be affected, but generally less severely, in terms of age at onset or prognosis, compared to the hemizygous males (i.e., with a single mutated copy of the gene). In some diseases, the presence of the mutations at the hemizygous state in males is even lethal, with the disease consequently being restricted to females. There is also an intermediate condition where only a small fraction of affected males are born. They usually have a profoundly deleterious disease with a very poor prognosis. All these possibilities will be illustrated in the following examples of X-linked EE.
Mutations in CDKL5/STK9 cause EE, preferentially in females
The CDKL5/STK9 gene was first incriminated in two unrelated girls with infantile spasm syndrome (ISSX2; 300672) associated with de novo balanced X-autosome translocations, t(X;7)(p22.3;p15) and t(X;6)(p22.3;q14), respectively. In both cases, the genomic rearrangements disrupted the CDKL5/STK9 gene. 14 Although CDKL5/STK9 was submitted to X inactivation in normal female somatic cells, the expressed protein was not functional in the two patients because of a preferential inactivation of the normal X. 14 Indeed, the inactivation of the translocated X might have diffused to the translocated autosome fragment, leading to a monosomy of the corresponding genomic region, which is lethal.
Mutations in CDKL5 were later on identified in female patients with atypical Rett syndrome. 15 These patients clearly had some Rett features such as deceleration of head growth, stereotypies and hand apraxia. In contrast, some signs were absent, such as a period of nearly normal development followed by regression with loss of acquired fine finger skill in early childhood and intensive eye communication, and the characteristic evolution of the Rett electroencephalogram. In the mouse brain, Weaving et al. (2004) showed that Cdkl5 expression overlapped with that of Mecp2 (OMIM: 300005), which is mutated in the classic Rett syndrome, 16 leading Tao et al. (2004) to postulate that these two genes may be involved in a common pathogenic pathway. 15
The characteristics of the epilepsy associated with CDKL5 mutations were defined by Bahi-Buisson and colleagues (2008), who showed in a retrospective study of 12 female patients with CDKL5 mutations that the epilepsy course had three successive stages: Stage I: early epilepsy (onset 1–10 weeks) with normal interictal EEG despite frequent convulsive seizures; Stage II: epileptic encephalopathy with infantile spasms and hypsarrhythmia; stage III: at the age of examination (3–19 years), seven patients were seizure free and six had developed refractory epilepsy with tonic seizures and myoclonia. 17 Moreover, early epilepsy with normal interictal EEG (stage I) and severe hypotonia seem to be key clinical features in identifying patients with CDKL5 mutations. 18
Epileptic encephalopathies as a part of the phenotypic spectrum associated with ARX mutations
Mutations in ARX were first identified in patients with three different phenotypes, including X-linked lissencephaly with abnormal genitalia (XLAG; MIM# 300215) 19, non syndromic X-linked mental retardation (MRX), 20 and X-linked infantile spasms (ISSX; MIM# 308350). 21 The phenotypic spectrum associated with ARX mutations includes severe EEs with or without brain malformations. Shoudbrige and colleagues (2010) recently reviewed families with ARX mutations reported in the literature. 22 Among the 97 families identified, Ohtahara syndrome, X-linked myoclonic epilepsy and X-linked infantile spasms (West syndrome) were reported in 1, 1 and 12 families, respectively. Other syndromes including seizures were also identified, including Partington syndrome (intellectual disability with dystonic movements, ataxia, and seizures), mental retardation with tonic seizures with dystonia, and infantile epileptic-dyskinetic encephalopathy, in 7, 1 and 4 families, respectively.
The ARX gene, which contains five coding exons (GenBank: NM_139058.2) (MIM] 300382), maps to Xp22 and encodes a 562 amino acid mature protein, which is expressed predominately in the fetal and adult brain, testis, skeletal muscle, and pancreas. 23–25 ARX is a paired-type homeobox protein that contains four polyalanine (PolyA) tracts, a homeodomain, and a conserved C-terminal aristaless domain. Male ARX-deficient mice have abnormal differentiation and deficient tangential migration of GABAergic (γ aminobutyric acid) interneurons in the ganglionic eminence, neocortex, and hippocampus, and abnormal testicular differentiation. 19
All types of mutations, missense, non-sense, insertions and deletions, have been reported in ARX. It is interesting to note that a phenotype-genotype correlation has recently emerged: 22, 26 non malformative syndromes including EE 27 are mostly due to expansions of polyalanine tracts located in the N terminal fragment of the protein whereas syndromes with brain malformations are mostly associated with mutations leading to a premature termination codon (PTC) (non-sense or splice mutations) or deletions encompassing several exons. These mutations lead to a loss of function by haploinsufficiency since the mRNAs with PTC are mainly destroyed in the cells by the non-sense-mediated mRNA decay (NMD) system. 28 Very recently, Kato et al. (2010) reported two families, comprising six males with Ohtahara syndrome in two generations, in which frameshift mutations in the terminal exon of the ARX gene, Ala524fsX534 and E536fsX672, were identified. It is noticeable that the PTC mutations localized in the last exon of genes escape the NMD system and most often cause the synthesis of a truncated protein. Interestingly, two patients developed West syndrome, and one of these later developed Lennox-Gastaut syndrome. The authors concluded that the analysis of ARX should be considered in sporadic or familial male patients with Ohtahara syndrome. 29
SRPX2 is responsible for rolandic seizures with speech apraxia and mental retardation
Roll et al. (2006) reported a 3-generation French family with oral and speech dyspraxia (OSP), rolandic seizures (RS), and mental retardation. Eight individuals had OSP, rolandic seizures and mental retardation and three had only OSP and mental retardation without seizures. Epileptic patients had the electroencephalographic hallmark of RS (centro-temporal spikes that tend to occur in clusters and are strongly activated during sleep) during the active phase of the epileptic seizures. 30 A whole-genome scan was performed and the gene was mapped to chromosome Xq. In the 20 cM large candidate interval, a missense mutation, N327S, was identified in the SRPX2 gene. SRPX2 is a secreted sushi-repeat-containing protein expressed in neurons of the human adult brain, including the rolandic area. The disease-causing mutation resulted in gain of glycosylation of the secreted mutant protein. 30 Roll at al. (2006) also described a 12-year-old boy who presented with focal seizures beginning with numbness of the fingers of the right hand and leg and a sudden fall. Neurologic examination revealed clonus at both knees and generalized hyperreflexia with an equivocal right plantar response. Neuropsychologic examination at age 15 years showed low average to average intellect with weakness in mathematical ability. His EEG showed left centrotemporal epileptiform activity with a rolandic horizontal dipole. MRI showed bilateral posterior perisylvian polymicrogyria (MIM # 300388), more severe on the left and extending back to involve the parieto-occipital regions. Two of his maternal aunts had mild mental retardation, and his mother and another aunt were of normal intelligence. In this family, a second mutation was identified within the first sushi domain of SRPX2. These data suggest that SRPX2 may play a role in the development and/or function of the perisylvian region critical for language and cognitive development. Interestingly, the orthologous Srpx2 gene is not expressed during murine embryogenesis suggesting that this gene is involved in the development and functioning of language areas in human cortex.30
A GENETICALLY-DETERMINED EPILEPTIC ENCEPHALOPATHY: DRAVET SYNDROME (OR SEVERE MYOCLONIC EPILEPSY OF INFANCY)
In contrast to the previously described EE, a large proportion of patients with Dravet syndrome have mutations in known genes, leading to their systematic testing if the clinical context is suggestive.
Clinical context
Dravet syndrome (DS), previously called severe myoclonic epilepsy of infancy (SMEI), is an intractable epileptic syndrome characterized by onset of seizures during the first year of life in a child with normal psychomotor development and no brain damage (MRI is normal at onset). Seizures are clonic or tonic-clonic and may be generalized or unilateral, with either side of the body being involved. They are mainly febrile and often prolonged, resulting in convulsive status epilepticus. Subsequently, children present myoclonic jerks, absences and focal seizures. Psychomotor development is delayed from the second year of life and ataxia may appear. Although these seizures are severe, the interictal EEGs remain free of spikes during the first years of the disorder. However, generalized spike waves with photosensitivity and focal abnormalities occur later on. 31
De novo mutations in SCN1A are the main cause of Dravet syndrome
A common genetic predisposition between SMEI and febrile seizures was first suggested by Benlounis et al. in 2001. 32 Furthermore, Singh et al., (2001) considered SMEI to be a severe phenotype of the “generalised epilepsy and febrile seizures plus” (GEFS+) familial context, particularly because of the presence of patients with SMEI in families with GEFS+ (MIM 604233). 33 GEFS+ is a variable autosomal dominant epileptic condition that also associates febrile and afebrile seizures. Affected family members present with phenotypes ranging from isolated febrile seizures to various idiopathic generalized epilepsy subtypes (epilepsy with grand mal seizures, childhood absence or juvenile myoclonic epilepsy) or can remain asymptomatic. The outcome is usually benign and patients are sensitive to classical antiepileptic treatments. 34 However, family members can occasionally experience focal seizures, be severely affected and/or pharmacoresistant, and even present with DS. 33 This common genetic background was confirmed by Claes and coworkers (2001) who identified seven de novo mutations in SCN1A, which had previously been incriminated in GEFS+,35 in 7 sporadic cases of SMEI. 36 The vast majority (6/7) of these mutations led to premature termination codon. 36 More recent studies have confirmed the high frequency of mutations of SCN1A in sporadic SMEI. 37–42
Different types of mutation in SCN1A cause Dravet syndrome
All types of mutations have been identified in the coding sequence of the SCN1A gene in patients with Dravet syndrome: missense, nonsense, and splice-site mutations, small deletions and insertions. The mutations are located all along the SCN1A gene. Missense mutations are the most common mutation type identified (about 40%). 42 The main remaining mutation types (nonsense, splice-site mutations, frameshift) introduce PTCs into the mRNA, which are probably recognized and degraded via the NMD system. This hypothesis is compatible with the effects of the other mutation types detected, including alteration of the initiation codon and the microdeletions that delete essential regions of the proteins, introduce PTC, or delete the whole gene at the heterozygous state. Indeed, during the past few years, many techniques have been developed in order to detect genomic rearrangements such as deletions or duplications: quantitative PCR (QPCR), 43 multiplex ligation-dependent probe amplification (MLPA) 44 and microarrays. 45 The analysis of series of patients without point mutations with these methods has identified several heterozygous rearrangements in SCN1A, variable in length and breakpoints and encompassing from a single exon to the whole gene. 42, 46, 47 Deletion of the whole gene was the most common rearrangement found, supporting haploinsufficiency as the main molecular mechanism responsible for Dravet syndrome. Analysis of the parents showed that most deletions occurred de novo. The nearby genes comprised in the deletion, including other genes encoding voltage-gated sodium channels (SCN7A and SCN9A), were different between patients, without evident variability of the phenotype. 42 However, patients with very large deletions could have consistent dysmorphic features including ear abnormalities, microcephaly, micrognathia and brachysyndactyly, likely related to the size of deletion and deletions of other genes than SCN1A. 48
In spite of many functional studies, it remains unclear how missense mutations can cause a clinical phenotype indistinguishable from that of PTC mutations or whole-gene deletion and why some missense mutations are associated with a severe phenotype, such as Dravet syndrome, and others with curable epilepsies as in GEFS+ families. Kanai et al. (2004) suggested a preferential location of the missense mutations leading to Dravet syndrome in the S5 and S6 segments and the S5–S6 intracellular boucle, which together form the “pore” of the Nav1.1 channel.21. 49 Depienne and colleagues (2009) reported that most missense .mutations causing Dravet syndrome affected highly conserved amino acids located in ion-transport sequences and resulted in chemically dissimilar changes in amino acid classes. However, these mutations were not preferentially located in S5–S6 segments, in contrast to the previous report (Figure 2). 42
The proportion of patients with SCN1A mutations is highly variable in reported studies, ranging from 33% 39, 50 to 80–100%.40, 51, 52 These discrepancies may be due to the sizes of the series, the methods of screening, and use of different clinical criteria to define Dravet syndrome. Supporting this hypothesis, the proportion of positive patients is higher in SMEI using strict criteria than in epileptic syndromes closely related to SMEI including ICEGTC (intractable childhood epilepsy with generalised tonic–clonic seizures) or SMEB (borderline SMEI). 51, 53 Considering only the patients with typical Dravet syndrome, approximately 20% of them do not have mutations in SCN1A, even when microrearrangements have been excluded. 41, 42 In addition, recent studies have established that rare pathogenic mutations in SCN1A, some of which are de novo, can also be identified in other infantile EEs such as cryptogenic generalized epilepsies, cryptogenic focal epilepsies or infantile spasms.50, 53 54 This extension of the clinical spectrum related to SCN1A has called into question the initial concept of SMEI, because neither clinical nor genetic criteria are sufficient to delimit accurately the various syndromes.
Dravet syndrome in a familial context of epilepsy
Few patients with DS have a parent or relative with a milder epileptic phenotype. Depienne and colleagues (2010) showed that mutations inherited from an asymptomatic or mildly affected parent were identified in 10% of the DS patients. Mosaicism (the mutation is present only in a fraction of germinal [germinal mosaicism] or non-germinal [somatic mosaicism] cells) was the main event associated with inherited SCN1A mutations in DS patients (about 70%). Parental mosaicism in DS was not a rare situation since it was found in at least 7% of families with an SCN1A mutation. When the level of somatic mosaicism was high, the parent could present with seizures, although he/she was less severely affected than his/her child who carried the mutation in all cells. The clinical status of the mosaic parent appeared to be somehow correlated with the amount of the mutation in his/her blood cells although this correlation was not strict. 55
Nevertheless, mosaicism is not the only situation accounting for inherited mutations as DS can also be encountered in the context of GEFS+ families. In that case, SCN1A missense mutations segregating in the family are associated with a wide phenotypic variability, with DS at the severe end of the spectrum. 33 Distinguishing mosaicism from de novo constitutional mutations or other situations is of particular concern in disorders that are frequently sporadic, such as DS, in order to give appropriate genetic counseling. 56 The risk of recurrence is, therefore, not null when the mutation is apparently de novo, which should be taken into account for genetic counseling.
A question that remains is whether SCN1A neomutations causing DS act through the same pathophysiological mechanisms as mutations found in GEFS+. Basically, mutations associated with clear loss-of-function are always associated with DS, except in one family recently described by Suls et al. (2010), in which a microdeletion of SCN1A segregated with a very variable phenotype in a 4-generation family. 57 In mosaic patients, the mutations with a loss-of-function effect can be associated with milder phenotypes reminiscent of those seen in GEFS+ if present in only some neurons. Conversely, missense mutations found in GEFS+ are generally associated with mild epileptic phenotypes but can occasionally cause DS. From a mechanistic point of view, it is likely that the pathophysiological pathways are different: de novo mutations would be sufficient to cause DS whereas missense mutations associated with GEFS+ would not, and additional genetic or non-genetic factors would be necessary to cause DS in the latter case.
Dravet-like syndrome in females associated with mutations in PCDH19 gene
At least 20% of patients with Dravet syndrome are negative for mutations or rearrangements in SCN1A. In addition, rare mutations in GABRG2 have been described in cases of SMEI belonging to GEFS+ families 58 but were not demonstrated in 29 sporadic cases. 38 These findings suggest that Dravet syndrome could be genetically heterogeneous (i.e. other genes are involved in DS).
Mutations in PCDH19 mainly affect females with a Dravet-like syndrome
In order to identify new genes responsible for the disorder in SCN1A mutation-negative patients, Depienne and colleagues screened 41 patients with DS for micro-rearrangements with high-density SNP microarrays. Interestingly, a hemizygous deletion on chromosome Xq22.1, encompassing the PCDH19 gene, was identified in one male patient. To confirm that PCDH19 was responsible for a Dravet-like syndrome, its coding region was sequenced in 73 additional SCN1A mutation-negative patients. Different point mutations (four missense and five truncating mutations) were identified in 11 unrelated female patients (15%). The spectrum of mutations includes non-sense mutations, small deletions/insertions introducing a frameshift as well as missense mutations affecting highly conserved amino acids in the protein, predominantly in the extracellular domain, which is presumably involved in cell-cell interaction. These mutations are therefore predicted to result in a loss-of-function of the mutated allele. 59
Protocadherin 19 is a 1148 amino acid transmembrane protein belonging to the protocadherin delta2 subclass of the cadherin superfamily, which is highly expressed in neural tissues and at different developmental stages. 60–63 The precise functions of the protein remain so far unknown. However, Delta protocadherins were reported to mediate cell-cell adhesion in vitro and cell sorting in vivo, and could regulate the establishment of neuronal connections during brain development. 64, 65
Patients with PCDH19 mutations could present with clinical features similar to patients with SCN1A mutations, including the association of early febrile and afebrile seizures, seizures occurring in clusters, developmental and language delays, behavioral disturbances, and cognitive regression. There were, however, slight but constant differences in the evolution of epilepsy, including fewer polymorphic seizures (in particular rare myoclonic jerks and atypical absences) in patients with PCDH19 mutations. These results show that PCDH19 plays a major role in epileptic encephalopathies, with a clinical spectrum overlapping that of DS. 59
PCDH19-linked epilepsies: an unusual mode of inheritance
Mutations in PCDH19 were first reported to cause EFMR (epilepsy and mental retardation limited to females). The clinical features of EFMR, unlike those of DS, are highly variable, even in members of the same family: onset of seizures is between 6 and 36 months and affected females present with a combination of febrile and afebrile seizures of various types and a variable degree of psychomotor delay and cognitive impairment, ranging from mild to severe mental retardation. 66 Dibbens et al. reported PCDH19 mutations in six large families and one small family with two affected sib pairs. 60 All the patients were familial cases that were, for the most part, already adults at the time of examination, and appeared socially integrated in that most of them were married and had children.
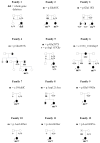
PCDH19-related epileptic encephalopathy therefore mainly affects females. In a large series of PCDH19 mutation-positive index cases in whom inheritance could be assessed, half of the mutations occurred de novo and half were inherited from fathers who were healthy, had no cognitive impairment and had never had febrile seizures or epilepsy. 59 (Depienne at al., personal communication) This heredity is very different from the X-linked mode of inheritance encountered for ARX or CDKL5 mutations since in this case only the heterozygous females are affected whereas the hemizygous males are asymptomatic and spared (Figure 3).
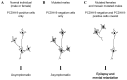
Several mechanisms have been suggested to account for the unusual mode of inheritance observed in PCDH19-linked epilepsy, one of which is cellular interference, a mechanism reminiscent of metabolic interference. 60 67, 68 This concept postulates that random inactivation of one X chromosome in mutated females generates tissue mosaicism (i.e., coexistence of PCDH19-positive an PCDH19-negative cells), which would be pathogenic by altering cell-cell interactions; normal individuals and mutated males, who are homogeneous for PCDH19-positive or PCDH19-negative cells, respectively, would not develop the disease (Figure 4). The identification of an affected male who was mosaic for the PCDH19 deletion in his fibroblasts, and therefore had PCDH19-positive and PCDH19-negative cells in this tissue, strongly supports the hypothesis of cellular interference as the main pathogenic mechanism associated with PCDH19 mutations. 59
PROSPECTS AND CONCLUSION
Identification of a mutation leads to genetic counseling
In the vast majority of EEs, genetic causes are very rare. However, after having excluded the most common, non-genetic causes, it is crucial to perform appropriate genetic analyses depending on the detailed phenotype of the patient, even if they are time-consuming and costly, since the identification of a mutation raises the possibility of a risk of recurrence in the family. A scrupulous reconstruction of the familial history has to be made and a pedigree drawn in order to determine the most probable mode of inheritance.
For AR diseases like EIEE3 associated with CG1 mutations, recurrence in the family is high (25%) and genetic counseling should be provided. The mutation(s) must firstly be identified in the index case. If the parents are related, we would expect to identify a mutation at the homozygous state. The analysis of the parents’ DNA is important to be able to conclude whether the mutation is on both copies of the gene in the affected child. However, in some rare cases only one parent carries the identified mutation, the other having a deletion encompassing the exon bearing the mutation in his/her spouse. If the parents are not related, patients most often have two different mutations in the CG1 gene; they are then called “compound heterozygous”. The study of the parents’ DNA is also used to exclude the possibility that both mutations are on the same copy of the gene. This needs to be verified before a prenatal diagnosis can be proposed.
Several genetically determined EEs are due to de novo mutations, including Dravet syndrome associated with mutations in SCN1A or PCDH19 and early infantile EE due to mutations in CDKL5/STK9 or STXBP1. Genetic counseling in these cases is a very delicate issue since the possibility of a germinal mosaicism has to be taken into account. Germinal mosaicism may be suspected in families in certain contexts: i) the mutation can be detected at a low level in the blood cells’ DNA of one parent (the mutation can be detected by direct sequencing when its amount is greater than 20% in DNA from blood cells); or ii) at least two children are affected and carry the same mutation, even if the mutation is not detected in the parent’s blood. Importantly, germinal mosaicism or somatic mosaicism undetectable from the blood may be missed in the absence of a second affected child, which means that the possibility of mosaicism should be consider for every sporadic de novo case. The frequency of mosaicism in Dravet syndrome was evaluated at 7% in a large cohort of patients with SCN1A mutations, but this might only be the tip of the iceberg. 55 The risk of recurrence is therefore not null and is difficult to evaluate when the mutation is apparently de novo, which raises the question of prenatal diagnosis in some cases.
The last condition most often concerns autosomal dominant diseases with phenotype variability and is illustrated by the occurrence of DS in families with GEFS+. In this case, the parent with a mild and benign phenotype transmits a SCN1A missense mutation to his/her child, who then presents with severe and intractable epilepsy and mental retardation. The occurrence of a DS phenotype has to be related to this mutation but its severity is likely determined by other, as yet unidentified factors, often called modifying factors, which can be of an environmental or genetic nature. Until these factors can be identified, the question of the recurrence of this severe phenotype must remain open.
Recent strategies to identify new genes responsible for epileptic encephalopathies
We are only just beginning to appreciate the importance of genetic EE and the consequences of the identification of a mutation in a family in terms of diagnosis and genetic counseling. From a fundamental point of view, the identification of a new gene leads to the deciphering of a new pathological mechanism and may open new therapeutic avenues. Advances in human molecular genetics are revealing the great genetic heterogeneity of infantile epileptic encephalopathies and the great variability of their pathological pathways. Moreover, one may suspect that the genes already identified represent only a small fraction of the genes involved in these disorders that can be considered as development disorders. Identifying these genes remains challenging. Nevertheless, the recent “technological leap” due to the development of next-generation sequencing, allowing the sequencing of the whole genome of coding sequences (exome) of a genome, opens new perspectives not only for monogenic56a but also polygenic forms of epileptic encephalopathies.
Identifying new genes responsible for monogenic forms of epileptic encephalopathies
Identifying genes in sporadic epileptic encephalopathies is difficult since the classical genetic strategies based on the study of large families are not possible. However, alternative genetic approaches are available; they are various and related to the context of the disease:
- As for ARX, CDKL5/STK9 and SRPX2, EE can be part of a complex phenotype with or without brain abnormalities. The strategy consisting in the screening of the responsible gene in series of patients with isolated EE of the same type can be fruitful. Some patients with isolated defined EEs, even if they were rare, had mutations in these genes.
- The study of GEFS+ and Dravet syndrome highlights the possibility of a gene identification strategy based on the detailed description of syndromes: the occurrence of Dravet syndrome in families with GEFS+ and the high sensitivity to fever in both conditions led to Dravet syndrome being considered as a part of the GEFS+ phenotypic spectrum. Claes et al. (2001) to be homogeneous therefore tested SCN1A, which had firstly been incriminated in GEFS+, as a candidate gene in sporadic patients with DS and identified the first de novo mutations of SCN1A. 36
- Genomic rearrangements have been identified in most of the genes known to be responsible for infantile EE. Comparative genomic hybridization (CGH) and SNP microarrays have enabled the entire genome to be screened at various resolutions. The detection of a de novo rearrangement in a patient points to a candidate region in which the causative gene is localized. The candidate region often contains several genes and the involvement of one particular gene needs to be confirmed by the identification of point mutations located in the coding region of this gene in patients with the same phenotype. As we saw in previous sections, this strategy was applied to identify the SPTAN1 and CDKL5/STK9 genes. When the rearrangement is inherited from an asymptomatic parent, its causality in the patient’s phenotype is more difficult to prove, as it may correspond to a benign copy number variant (CNV) or copy number polymorphism (CNP). However, it cannot be excluded that a recessive mutation in a gene of the region has been transmitted on the undeleted remaining copy by the other parent. Association of an inherited CNV with a phenotype in more than one patient would argue in a favor of such a recessive gene and lead to the sequencing of candidate genes in the regions concerned.
Identifying susceptibility genes in polygenic-determined epileptic encephalopathies
Since the vast majority of cases of EE are sporadic, the most frequent inheritance of EE is usually thought of as being complex or multifactorial, resulting from the interaction between several genes (susceptibility genes) and environmental factors. The identification of these genes requires non-parametric analyses (genetic analyses that do not depend on parameters of monogenic diseases as the mode of inheritance, penetrance, rate of phenocopies, etc.). For these analyses, several conditions are needed: i) a large number of affected individuals (at least in the hundreds) and a similar number of controls well-matched for age, sex and ethnic background, ii) a previous exclusion of monogenic forms, and iii) a low genetic heterogeneity of the population (a great genetic heterogeneity could otherwise mask or dilute the effect of individual susceptibility genes). These conditions are difficult to obtain in EEs, which are rare diseases affecting children (problem of constituting a control group) with a well-known etiological heterogeneity (Table 1). However, such a strategy has already been performed, in which the entities Landau-Kleffner syndrome (LKS), continuous spike-and-waves during sleep syndrome (CSWS), and benign childhood epilepsy with centrotemporal spikes (BCECTS) and benign rolandic epilepsy, were considered to be part of a single continuous spectrum of disorders. 69 The centrotemporal spikes (CTS) were considered to be an endophenotype determined by a common set of susceptibility genes, the different phenotypes being determined by other susceptibility genes and/or environmental factors. A genome-wide study showed a linkage of the CTS endophenotype to 11p13, especially with polymorphic markers in the ELP4 (Elongator Protein Complex 4) gene. 70 The yeast Elp4 protein is one of the 6 subunits of Elongator. The Elongator complex is involved in transcription of several genes that regulate the actin cytoskeleton, cell motility and migration. These functions are crucial in the nervous system for nerve cell growth cone motility, axon outgrowth and guidance, neuritogenesis and neuronal migration during development. 71, 72 This gene is a good candidate-by-function. It remains to identify functional variants in the vicinity of the ELP4 gene to confirm its genetic involvement. This strategy could be applied to other clinical or EEG traits.
In conclusion, in the past ten years, a great amount of genetic data has accumulated, updating our knowledge on the etiologies and pathophysiological mechanisms of infantile epileptic encephalopathies. We can expect this chapter to become incomplete in the very next future and hope that all this basic science will open soon new therapeutic avenues to cure these devastating disorders.
Acknowledgments
Our research was supported financially by INSERM, AP-HP and UPMC-P6.
References
- 1.
- Dulac O. Epileptic encephalopathy. Epilepsia. 2001;42(Suppl 3):23–26. [PubMed: 11520318]
- 2.
- Nabbout R, Dulac O. Epileptic encephalopathies: a brief overview. J Clin Neurophysiol. 2003 11–12;20(6):393–397. [PubMed: 14734929]
- 3.
- Guerrini R. Epilepsy in children. Lancet. 2006 Feb 11;367(9509):499–524. [PubMed: 16473127]
- 4.
- Gerber SH, Rah JC, Min SW, et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008 Sep 12;321(5895):1507–1510. [PMC free article: PMC3235364] [PubMed: 18703708]
- 5.
- Verhage M, Maia AS, Plomp JJ, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000 Feb 4;287(5454):864–869. [PubMed: 10657302]
- 6.
- Saitsu H, Kato M, Mizuguchi T, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008 Jun;40(6):782–788. [PubMed: 18469812]
- 7.
- Hamdan FF, Piton A, Gauthier J, et al. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol. 2009 Jun;65(6):748–753. [PubMed: 19557857]
- 8.
- Tohyama J, Akasaka N, Osaka H, et al. Early onset West syndrome with cerebral hypomyelination and reduced cerebral white matter. Brain Dev. 2008 May;30(5):349–355. [PubMed: 18065176]
- 9.
- Saitsu H, Tohyama J, Kumada T, et al. Dominant-negative mutations in alpha-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am J Hum Genet. Jun 11;86(6):881–891. [PMC free article: PMC3032058] [PubMed: 20493457]
- 10.
- Metral S, Machnicka B, Bigot S, Colin Y, Dhermy D, Lecomte MC. AlphaII-spectrin is critical for cell adhesion and cell cycle. J Biol Chem. 2009 Jan 23;284(4):2409–2418. [PubMed: 18978357]
- 11.
- Molinari F, Raas-Rothschild A, Rio M, et al. Impaired mitochondrial glutamate transport in autosomal recessive neonatal myoclonic epilepsy. Am J Hum Genet. 2005 Feb;76(2):334–339. [PMC free article: PMC1196378] [PubMed: 15592994]
- 12.
- Molinari F, Kaminska A, Fiermonte G, et al. Mutations in the mitochondrial glutamate carrier SLC25A22 in neonatal epileptic encephalopathy with suppression bursts. Clin Genet. 2009 Aug;76(2):188–194. [PubMed: 19780765]
- 13.
- Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE. Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem. 2002 May 31;277(22):19289–19294. [PubMed: 11897791]
- 14.
- Kalscheuer VM, Tao J, Donnelly A, et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet. 2003 Jun;72(6):1401–1411. [PMC free article: PMC1180301] [PubMed: 12736870]
- 15.
- Tao J, Van Esch H, Hagedorn-Greiwe M, et al. Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet. 2004 Dec;75(6):1149–1154. [PMC free article: PMC1182152] [PubMed: 15499549]
- 16.
- Weaving LS, Christodoulou J, Williamson SL, et al. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet. 2004 Dec;75(6):1079–1093. [PMC free article: PMC1182143] [PubMed: 15492925]
- 17.
- Bahi-Buisson N, Kaminska A, Boddaert N, et al. The three stages of epilepsy in patients with CDKL5 mutations. Epilepsia. 2008 Jun;49(6):1027–1037. [PubMed: 18266744]
- 18.
- Bahi-Buisson N, Nectoux J, Rosas-Vargas H, et al. Key clinical features to identify girls with CDKL5 mutations. Brain. 2008 Oct;131(Pt 10):2647–2661. [PubMed: 18790821]
- 19.
- Kitamura K, Yanazawa M, Sugiyama N, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002 Nov;32(3):359–369. [PubMed: 12379852]
- 20.
- Bienvenu T, Poirier K, Friocourt G, et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet. 2002 Apr 15;11(8):981–991. [PubMed: 11971879]
- 21.
- Stromme P, Mangelsdorf ME, Shaw MA, et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet. 2002 Apr;30(4):441–445. [PubMed: 11889467]
- 22.
- Shoubridge C, Fullston T, Gecz J. ARX spectrum disorders: making inroads into the molecular pathology. Hum Mutat. Aug;31(8):889–900. [PubMed: 20506206]
- 23.
- Miura H, Yanazawa M, Kato K, Kitamura K. Expression of a novel aristaless related homeobox gene ‘Arx’ in the vertebrate telencephalon, diencephalon and floor plate. Mech Dev. 1997 Jul;65(1–2):99–109. [PubMed: 9256348]
- 24.
- Collombat P, Mansouri A, Hecksher-Sorensen J, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003 Oct 15;17(20):2591–2603. [PMC free article: PMC218152] [PubMed: 14561778]
- 25.
- Gecz J, Cloosterman D, Partington M. ARX: a gene for all seasons. Curr Opin Genet Dev. 2006 Jun;16(3):308–316. [PubMed: 16650978]
- 26.
- Kato M, Saitoh S, Kamei A, et al. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome). Am J Hum Genet. 2007 Aug;81(2):361–366. [PMC free article: PMC1950814] [PubMed: 17668384]
- 27.
- Guerrini R, Moro F, Kato M, et al. Expansion of the first PolyA tract of ARX causes infantile spasms and status dystonicus. Neurology. 2007 Jul 31;69(5):427–433. [PubMed: 17664401]
- 28.
- Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008 Jul;18(7):315–321. [PubMed: 18524595]
- 29.
- Kato M, Koyama N, Ohta M, Miura K, Hayasaka K. Frameshift mutations of the ARX gene in familial Ohtahara syndrome. Epilepsia. Apr 2; [PubMed: 20384723]
- 30.
- Roll P, Rudolf G, Pereira S, et al. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006 Apr 1;15(7):1195–1207. [PubMed: 16497722]
- 31.
- Dravet C, Catani C, Bureau M, Roger J. Partial epilepsies in infancy: a study of 40 cases. Epilepsia. 1989 11–12;30(6):807–812. [PubMed: 2512116]
- 32.
- Benlounis A, Nabbout R, Feingold J, et al. Genetic predisposition to severe myoclonic epilepsy in infancy. Epilepsia. 2001 Feb;42(2):204–209. [PubMed: 11240590]
- 33.
- Singh R, Andermann E, Whitehouse WP, et al. Severe myoclonic epilepsy of infancy: extended spectrum of GEFS+ Epilepsia. 2001 Jul;42(7):837–844. [PubMed: 11488881]
- 34.
- Scheffer IE, Berkovic SF. Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain. 1997 Mar;120(Pt 3):479–490. [PubMed: 9126059]
- 35.
- Escayg A, Goldin AL. Sodium channel SCN1A and epilepsy: Mutations and mechanisms. Epilepsia. May 28; [PMC free article: PMC2937162] [PubMed: 20831750]
- 36.
- Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001 Jun;68(6):1327–1332. [PMC free article: PMC1226119] [PubMed: 11359211]
- 37.
- Sugawara T, Mazaki-Miyazaki E, Fukushima K, et al. Frequent mutations of SCN1A in severe myoclonic epilepsy in infancy. Neurology. 2002 Apr 9;58(7):1122–1124. [PubMed: 11940708]
- 38.
- Ohmori I, Ouchida M, Ohtsuka Y, Oka E, Shimizu K. Significant correlation of the SCN1A mutations and severe myoclonic epilepsy in infancy. Biochem Biophys Res Commun. 2002 Jul 5;295(1):17–23. [PubMed: 12083760]
- 39.
- Nabbout R, Gennaro E, Dalla Bernardina B, et al. Spectrum of SCN1A mutations in severe myoclonic epilepsy of infancy. Neurology. 2003 Jun 24;60(12):1961–1967. [PubMed: 12821740]
- 40.
- Claes L, Ceulemans B, Audenaert D, et al. De novo SCN1A mutations are a major cause of severe myoclonic epilepsy of infancy. Hum Mutat. 2003 Jun;21(6):615–621. [PubMed: 12754708]
- 41.
- Marini C, Mei D, Temudo T, et al. Idiopathic epilepsies with seizures precipitated by fever and SCN1A abnormalities. Epilepsia. 2007 Sep;48(9):1678–1685. [PubMed: 17561957]
- 42.
- Depienne C, Trouillard O, Saint-Martin C, et al. Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. J Med Genet. 2009 Mar;46(3):183–191. [PubMed: 18930999]
- 43.
- Laccone F, Junemann I, Whatley S, et al. Large deletions of the MECP2 gene detected by gene dosage analysis in patients with Rett syndrome. Hum Mutat. 2004 Mar;23(3):234–244. [PubMed: 14974082]
- 44.
- Montagna M, Dalla Palma M, Menin C, et al. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003 May 1;12(9):1055–1061. [PubMed: 12700174]
- 45.
- Bignell GR, Huang J, Greshock J, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004 Feb;14(2):287–295. [PMC free article: PMC327104] [PubMed: 14762065]
- 46.
- Suls A, Claeys KG, Goossens D, et al. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Hum Mutat. 2006 Sep;27(9):914–920. [PubMed: 16865694]
- 47.
- Marini C, Scheffer IE, Nabbout R, et al. SCN1A duplications and deletions detected in Dravet syndrome: Implications for molecular diagnosis. Epilepsia. 2009 Mar 11; [PubMed: 19400878]
- 48.
- Davidsson J, Collin A, Olsson ME, Lundgren J, Soller M. Deletion of the SCN gene cluster on 2q24.4 is associated with severe epilepsy: an array-based genotype-phenotype correlation and a comprehensive review of previously published cases. Epilepsy Res. 2008 Sep;81(1):69–79. [PubMed: 18539002]
- 49.
- Kanai K, Hirose S, Oguni H, et al. Effect of localization of missense mutations in SCN1A on epilepsy phenotype severity. Neurology. 2004 Jul 27;63(2):329–334. [PubMed: 15277629]
- 50.
- Wallace RH, Hodgson BL, Grinton BE, et al. Sodium channel alpha1-subunit mutations in severe myoclonic epilepsy of infancy and infantile spasms. Neurology. 2003 Sep 23;61(6):765–769. [PubMed: 14504318]
- 51.
- Fujiwara T. Clinical spectrum of mutations in SCN1A gene: severe myoclonic epilepsy in infancy and related epilepsies. Epilepsy Res. 2006 Aug;70(Suppl 1):S223–230. [PubMed: 16806826]
- 52.
- Fukuma G, Oguni H, Shirasaka Y, et al. Mutations of neuronal voltage-gated Na+ channel alpha 1 subunit gene SCN1A in core severe myoclonic epilepsy in infancy (SMEI) and in borderline SMEI (SMEB). Epilepsia. 2004 Feb;45(2):140–148. [PubMed: 14738421]
- 53.
- Harkin LA, McMahon JM, Iona X, et al. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007 Mar;130(Pt 3):843–852. [PubMed: 17347258]
- 54.
- Zucca C, Redaelli F, Epifanio R, et al. Cryptogenic epileptic syndromes related to SCN1A: twelve novel mutations identified. Arch Neurol. 2008 Apr;65(4):489–494. [PubMed: 18413471]
- 55.
- Depienne C, Trouillard O, Gourfinkel-An I, et al. Mechanisms for variable expressivity of inherited SCN1A mutations causing Dravet syndrome. J Med Genet. Jun;47(6):404–410. [PubMed: 20522430]
- 56.
- Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002 Oct;3(10):748–758. [PubMed: 12360233]
- 56a.
- Veeramah KR, O'Brien JE, Meisler MH, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012 Mar;90(3):502–510. [PMC free article: PMC3309181] [PubMed: 22365152]
- 57.
- Suls A, Velizarova R, Yordanova I, et al. Four generations of epilepsy caused by an inherited microdeletion of the SCN1A gene. Neurology. Jul 6;75(1):72–76. [PubMed: 20484682]
- 58.
- Harkin LA, Bowser DN, Dibbens LM, et al. Truncation of the GABA(A)-receptor gamma2 subunit in a family with generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2002 Feb;70(2):530–536. [PMC free article: PMC384926] [PubMed: 11748509]
- 59.
- Depienne C, Bouteiller D, Keren B, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009 Feb;5(2):e1000381. [PMC free article: PMC2633044] [PubMed: 19214208]
- 60.
- Dibbens LM, Tarpey PS, Hynes K, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008 Jun;40(6):776–781. [PMC free article: PMC2756413] [PubMed: 18469813]
- 61.
- Hynes K, Tarpey P, Dibbens LM, et al. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J Med Genet. Mar;47(3):211–216. [PubMed: 19752159]
- 62.
- Gaitan Y, Bouchard M. Expression of the delta-protocadherin gene Pcdh19 in the developing mouse embryo. Gene Expr Patterns. 2006 Oct;6(8):893–899. [PubMed: 16682261]
- 63.
- Wolverton T, Lalande M. Identification and characterization of three members of a novel subclass of protocadherins. Genomics. 2001 Aug;76(1–3):66–72. [PubMed: 11549318]
- 64.
- Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 2005 Oct;17(5):446–452. [PubMed: 16099637]
- 65.
- Redies C, Vanhalst K, Roy F. delta-Protocadherins: unique structures and functions. Cell Mol Life Sci. 2005 Dec;62(23):2840–2852. [PubMed: 16261259]
- 66.
- Scheffer IE, Turner SJ, Dibbens LM, et al. Epilepsy and mental retardation limited to females: an under-recognized disorder. Brain. 2008 Apr;131(Pt 4):918–927. [PubMed: 18234694]
- 67.
- Wieland I, Jakubiczka S, Muschke P, et al. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet. 2004 Jun;74(6):1209–1215. [PMC free article: PMC1182084] [PubMed: 15124102]
- 68.
- Johnson WG. Metabolic interference and the + - heterozygote. a hypothetical form of simple inheritance which is neither dominant nor recessive. Am J Hum Genet. 1980 May;32(3):374–386. [PMC free article: PMC1686061] [PubMed: 6770678]
- 69.
- Rudolf G, Valenti MP, Hirsch E, Szepetowski P. From rolandic epilepsy to continuous spike-and-waves during sleep and Landau-Kleffner syndromes: insights into possible genetic factors. Epilepsia. 2009 Aug;50(Suppl 7):25–28. [PubMed: 19682046]
- 70.
- Strug LJ, Clarke T, Chiang T, et al. Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4). Eur J Hum Genet. 2009 Sep;17(9):1171–1181. [PMC free article: PMC2729813] [PubMed: 19172991]
- 71.
- Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006 Oct 6;24(1):139–148. [PubMed: 17018299]
- 72.
- Close P, Hawkes N, Cornez I, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell. 2006 May 19;22(4):521–531. [PubMed: 16713582]
- Review Genes of early-onset epileptic encephalopathies: from genotype to phenotype.[Pediatr Neurol. 2012]Review Genes of early-onset epileptic encephalopathies: from genotype to phenotype.Mastrangelo M, Leuzzi V. Pediatr Neurol. 2012 Jan; 46(1):24-31.
- Review Diagnostic Approach to Genetic Causes of Early-Onset Epileptic Encephalopathy.[J Child Neurol. 2016]Review Diagnostic Approach to Genetic Causes of Early-Onset Epileptic Encephalopathy.Gürsoy S, Erçal D. J Child Neurol. 2016 Mar; 31(4):523-32. Epub 2015 Aug 13.
- Review Dravet syndrome as part of the clinical and genetic spectrum of sodium channel epilepsies and encephalopathies.[Epilepsia. 2019]Review Dravet syndrome as part of the clinical and genetic spectrum of sodium channel epilepsies and encephalopathies.Mei D, Cetica V, Marini C, Guerrini R. Epilepsia. 2019 Dec; 60 Suppl 3:S2-S7.
- Review The Broad Clinical Spectrum of Epilepsies Associated With Protocadherin 19 Gene Mutation.[Front Neurol. 2021]Review The Broad Clinical Spectrum of Epilepsies Associated With Protocadherin 19 Gene Mutation.Dell'Isola GB, Vinti V, Fattorusso A, Tascini G, Mencaroni E, Di Cara G, Striano P, Verrotti A. Front Neurol. 2021; 12:780053. Epub 2022 Jan 17.
- Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females.[PLoS Genet. 2009]Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females.Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O, Benyahia B, Quelin C, Carpentier W, Julia S, et al. PLoS Genet. 2009 Feb; 5(2):e1000381. Epub 2009 Feb 13.
- Genes in infantile epileptic encephalopathies - Jasper's Basic Mechanisms of the...Genes in infantile epileptic encephalopathies - Jasper's Basic Mechanisms of the Epilepsies
- BBS5 Bardet-Biedl syndrome 5 [Homo sapiens]BBS5 Bardet-Biedl syndrome 5 [Homo sapiens]Gene ID:129880Gene
- Gene Links for GEO Profiles (Select 43163488) (1)Gene
Your browsing activity is empty.
Activity recording is turned off.
See more...