NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Introduction
Telomeres are DNA-protein complexes found at the ends of linear eukaryotic chromosomes. The telomeric complexes prevent the chromosome ends from being recognized and processed as double strand breaks (for a review, see ref. 1). In most eukaryotes, the telomeric DNA consists of short repeats (5–8 bp) containing G clusters and the complementary C clusters, as well as other short repeats that are interspersed with the G:C clusters. The total length of the telomeric DNA varies considerably among different organisms, ranging from about 0.3 kb in the ciliate Tetrahymena thermophila and in the yeast Saccharomyces cerevisiae, to 10–20 kb in humans, and up to 50 kb in the laboratory mouse Mus musculus. The telomeric G-rich strand runs in the 5' to 3' direction towards the termini of the chromosomes and ends as a short single-stranded 3'-overhang. The length of the overhang also varies, e.g., in Tetrahymena it consists of 12–16 nucleotide residues and in humans it contains 100–200 residues.1 Both the double-stranded and the single-stranded telomeric repeats are associated with specific proteins.2–4 The telomeric complex also includes additional proteins that do not directly bind the telomeric DNA (e.g., ref. 5).
Following the elucidation of the discontinuous mode of DNA replication, it has been realized that replication of the the ends of linear chromosomes cannot be completed by the normal replication apparatus.6,7 A search for another mechanism for chromosome end replication led to the discovery of the enzyme telomerase that specifically replicates telomeric DNA in most eukaryotic cells.8 Telomerase is a reverse transcriptase that utilizes, as a template, a sequence within an RNA molecule which is an integral subunit of the enzyme.9 In cells lacking telomerase, the telomeres shorten after each round of chromosome replication.10 Telomerase prevents the shortening by elongating the single-stranded, G-rich 3' telomeric overhang, thereby providing a template for synthesis of the complementary C-rich strand by the normal replication apparatus.11 It is also capable of de novo synthesis of telomeres by addition of telomeric sequences to chromosome fragments that do not end with telomeric repeats.12,13
Telomerase was first discovered in the ciliate Tetrahymena thermophila and was later identified in cells of many other eukaryotic species. Several recent reviews have discussed the properties of the ciliate, yeast and human telomerases.2,3,14–16 In this review, we shall briefly describe some of the more recent data on the structure and function of these enzymes, which indicate that they share many properties. However, we shall focus our discussion on the Tetrahymena telomerase. In particular, we shall discuss in detail the in vitro reaction of primer extension by the telomerase, which mimics telomere elongation by the enzyme in vivo.
Structure and Assembly of the Telomerase Ribonucleoprotein
Genetic and biochemical studies of telomeres and telomerase, performed primarily in yeast, indicated that the telomerase can associate with some of the DNA binding proteins found in the telomeric complex (reviewed in refs. 3,4). This large and dynamic complex also includes components of the normal replication apparatus that carry out the replication of the telomeric C-rich strand, in concert with the extension of the G-rich strand by the telomerase.4 Other proteins that are associated with the large telomeric complex include the Ku and the RAD50/MRE11/NSB1 subcomplexes, which are also known to be recruited to double strand breaks in DNA.3,4
However, the basic core of all telomerases characterized so far consists of two components—the catalytic protein subunit, designated Telomerase Reverse Transcriptase (TERT),17,18 and the integral telomerase RNA designated TER.19 The assembly of catalytically active core telomerases from the Tetrahymena TERT and TER, and from the corresponding human components has been accomplished in vitro.20–22 In both systems, these two components were synthesized in rabbit reticulocyte lysates and the assembly process required the presence of additional factors found in the lysates. In the human system these factors were identified as components of the hsp90 chaperone complex.23 The use of this reconstitution protocol allowed mapping of the regions of TER and TERT that are required for the assembly of the core enzyme, as described below.
The TER Subunit: Regions in the RNA That Are Involved in the Assembly of the Telomerase Ribonucleoprotein
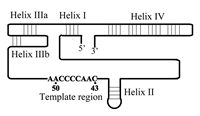
The TER subunit in each of the telomerases includes a short template sequence (3'-AACCCCAAC-5' in Tetrahymena and 3'-UCCCAAUC-5' in man) that encodes the telomeric repeats found in these species.9,24 However, other regions in the RNA also play essential roles in the telomerase ribonucleoprotein (RNP) assembly and in the catalytic activity of the enzyme. Identification of these regions was aided by phylogenetic studies of TER in various organisms. Ciliate telomerase RNAs from 12 species have been sequenced and found to range in length from 148 to 209 nucleotides. Comparison of the RNA sequences revealed that their secondary structures were conserved in evolution, even though their primary structures were not.25–27 Figure 1 shows a schematic drawing of one of these ciliate TER molecules, that of the Tetrahymena telomerase. This RNA consists of 159 nucleotide residues and includes the conserved helices (and the corresponding loops) designated I, II, IIIa, IIIb and IV, whose relative positions are indicated. It should be noted that helices IIIa and IIIb constitute a pseudoknot structure. The nine nucleotides constituting the template, which are also shown in Figure 1, are found within a region devoid of secondary structure.
The involvement of the various regions of TER in the assembly of the Tetrahymena telomerase RNP has been studied by genetic and RNA footprinting methods. Telomerase RNA mutants were prepared in vivo through transfection of Tetrahymena cells with high copy number plasmids encoding mutant RNAs. The enzymes were assembled in the transfected cells from the mutant RNAs and the endogeneous TERT. Minor amounts of wild-type enzyme that were also produced in the cells from RNA that was expressed from the endogeneous TER gene did not interfere with the analysis of the mutant enzymes.28,29 Telomerase RNA mutants were also prepared in vitro by two different enzyme reconstitution procedures. In one procedure, the RNA component in partially purified telomerase was inactivated by digestion with the enzyme micrococcal nuclease. Then, telomerase activity was reconstituted by addition of mutant RNAs that had been synthesized in vitro.30 In the second procedure, which has already been mentioned above, telomerase RNA and TERT were synthesized and assembled in rabbit reticulocyte lysates.20,22
The in vivo method of telomerase assembly revealed that disruption of the pseudoknot base pairing prevented the stable assembly of an active telomerase.31 The in vitro reconstitution provided more detailed information on the effects of various mutations in TER on the assembly process. It was found that regions in stem-loop II and two conserved nucleotides 5' of stem-loop II (CA nucleotides nos. 15–16) are required for binding of TER and TERT.32 Another study revealed that even though loop IV is not required for the initial binding of TER and TERT, it does play a role in a subsequent conformational change of the pseudoknot region, which is essential for enzyme activity.33
Recently, telomerase RNAs from 32 vertebrate species were sequenced and their lengths were found to range from 382 to 559 nucleotides. The secondary structures of these RNAs were also found to be conserved.34 A comparison between the vertebrate and the ciliate telomerase RNA structures indicated that the vertebrate RNAs contain elements that were also found in the ciliate RNAs. These common elements include some hairpin structures and the pseudoknot, as well as a the region devoid of secondary structure that contains the template sequence. In addition, the vertebrate telomerase RNAs contain sequence and secondary structure elements (H/ACA sequences) that bind proteins, such as dyskerin, which are also found in complexes containing small nucleolar RNAs (snoRNAs).35 These proteins are required for assembly of the vertebrate telomerase RNPs in vivo.35
The TERT Subunit: Regions in the Protein That Are Involved in the Assembly of the Telomerase Ribonucleoprotein Complex
Figure 2 shows a linear map of the Tetrahymena Telomerase TERT subunit that consists of 1117 amino acids.22,36,37 As indicated, this protein contains seven reverse transcriptase (RT) motifs designated 1, 2, A, B', C, D and E. These motifs were conserved in evolution among all reverse transcriptases.38 In particular, motifs A and C contain three conserved aspartate residues, which are found at the active sites of many reverse transcriptases and DNA polymerases. The region in TERT that contains the reverse transcriptase motifs is likely to constitute a separate domain having a structure that bears some resemblance to a human right hand, like the reverse transcriptases and DNA polymerases that have been structurally analyzed (for a review see ref. 39). Another region of the Tetrahymena TERT, which extends between amino acids 195 and 516 and does not overlap with the reverse transcriptase region, was found to be necessary and sufficient for specific binding of TER.37 As Figure 2 indicates, this region contains a motif designated T, which was conserved among all telomerases. It also contains other motifs, CP and CP2, that were conserved among ciliate telomerases (see refs. 37,40). Another motif shown in Figure 2 is T2, which maps closer to the N-terminus of TERT. T2 was conserved among all telomerases. Mutations in motifs CP and T were found to reduce the binding of TERT with TER. However, these reductions could not entirely account for the corresponding reductions in the catalytic activity of the telomerase. Based on these data, it was hypothesized that these motifs could be involved not only in the assembly of the core enzyme, but also in specific positioning or orientation of the RNA subunit in the telomerase RNP, which is required for optimal telomerase activity.36 On the other hand, it was found that the contribution of the RT motifs to the binding of TER and TERT was only marginal.36,37 These findings are compatible with a model assuming that TERT consists of at least two separate domains: a reverse transcriptase domain containing the RT motifs, which is only loosely associated with the template region of TER, and an RNA binding domain including the T, CP and CP2 motifs, which strongly binds other regions of TER. This model will be further discussed in section 3, in relation to the mechanism of primer extension by the telomerase.

Figure 2
A linear scheme of the TetrahymenaTelomerase Reverse Transcriptase (TERT) subunit. The letters and the numbers designate amino acid motifs discussed in the text. This scheme is a modified version of Figure 1A in ref. .
It should also be noted that in the human hTERT, which consists of 1132 amino acids, a region containing the N-terminal 617 amino acids was found to be necessary and sufficient for binding the human telomerase RNA. This region is also separate from the region containing the RT motifs.37,41
Telomerase Auxilliary Proteins and the Telomerase Holoenzyme
The structure of the telomerase holoenzyme has not yet been thoroughly characterized. However, it is interesting to note that the native Tetrahymena telomerase RNP may contain two auxiliary proteins designated p80 and p95 that copurify with the telomerase activity and the telomerase RNA.42 Human and mouse p80 homologues have been also isolated and found to associate with the human TERT and the mouse TERT, respectively, in vitro.43,44 Addition of the Tetrahymena and the human p80 homologue to the corresponding reconstituted telomerases did not affect the activities of these enzymes.22,45 However, in living Tetrahymena cells p80 and p95 appear to play a role in telomere metabolism.46 As mentioned above, the hsp90 chaperone complex is required for human telomerase assembly in vitro. Recently, it has been reported that some components of this complex remain associated with the human telomerase after the assembly process and might also play a role in the catalytic activities of the enzyme.47
A study of the yeast telomerase has indicated that it contains at least two functionally interacting RNA molecules that are simultaneously engaged in the extension of two primer DNA molecules.48 This finding indicated that the functional yeast telomerase is a dimer consisting of two active telomerase RNPs. Recent experiments have also indicated that the human telomerase can form dimers.49 At the present time, there is no evidence for dimeric ciliate telomerases.
Detailed Analysis of Primer Extension by the Telomerase
As discussed above, one function of telomerase is elongation of preexisting telomeric repeats and thereby prevention of shortening of telomeres due to the end replication problem. In vitro, this activity is mimicked by elongation of single-stranded DNA primers containing G-rich telomeric repeats, or of single-stranded telomeric 3' overhangs of double-stranded DNA molecules. A second function of telomerase is chromosome healing, a term used for de novo synthesis of telomeres at double strand breaks in chromosomal DNA. In ciliates, this function is required for adding telomeric repeats to DNA fragments generated by cleavage of the micronuclear chromosomes during the developmental process that leads to formation of macronuclei.12 In humans and other mammals, de novo telomere synthesis by telomerase may occur at ends generated by chromosome breakage events.13 The de novo synthesis of telomeres has been mimicked in vitro by extension of single-stranded non-telomeric oligonucleotides, or single stranded 3' extensions of double-stranded DNA that do not contain telomeric repeats. In this section, we shall first present a model for primer extension by telomerase based on earlier data. Then, we shall review the data that led to the formulation of this model and more recent data on the primer extension process. In particular, we shall discuss studies that contributed to the understanding of the special characteristics of telomerases vs. other reverse transcriptases, namely the exclusive use of an integral RNA template and the special mechanism employed for the synthesis of the short telomeric repeats. Finally, we shall present and discuss an updated model for primer extension by telomerase.
Model for the Telomerase-Catalysed Primer Extension Reaction
Most of the early in vitro studies of primer extension by the Tetrahymena telomerase were carried out with primers consisting entirely of G-rich telomeric repeats, or with primers containing a G-rich telomeric sequence at the 3'-terminus (for a review, see ref. 50). Both types of primers were extended in the presence of dTTP and dGTP, one of which was radioactively labeled. Then, the products were resolved by electrophoresis in a denaturing gel and were identified by autoradiography, or by use of a phosphorimager. Figure 3 shows typical gel profiles obtained in such assays. These profiles contain high-intensity bands that appear at intervals of six nucleotides. The periodicity is not visible at the top of the gel due to the poor resolution of the longer products. Each of the bands contains a fragment resulting from a relatively long pause in the elongation of the primers. Additional low-intensity bands are also visible between the strong bands. These bands contain fragments generated by shorter pauses that occurred after the addition of each successive nucleotide to the growing chains. Experiments of this type were carried out with primers whose ends were aligned at different positions along the RNA template region. These and other early experiments led to formulation of the model shown in Figure 4. According to this model, each primer is first aligned and forms a short double helix with the RNA template region in the telomerase. The alignment is determined by the particular repeat permutation found at the 3' terminus of the primer. Next, the primer is extended downstream in the direction 5' to 3'. The synthesis ends at the 5' terminus of the template region. The strong periodic pauses observed in the elongation of the primer occur at this stage. Subsequently, the Watson-Crick hydrogen bonds between the primer and the RNA are disrupted and the 3' end of the primer, along with the active site of the enzyme, is translocated to the 3' terminus of the template region. The translocation is followed by synthesis of the next telomeric repeat sequence, and so on.

Figure 4
Model for telomere repeat synthesis by the Tetrahymena telomerase. The 9 nucleotides constituting the primer region are shown. The protein subunit(s) are schematically depicted as an ellipse. Further details are presented in the text.
The Tetrahymena telomerase was found to be processive—the primers were elongated by 520 nucleotides before half of the enzyme molecules dissociated from the DNA products.51 This rather high processivity of the enzyme led to the suggestion that, in addition to the active site, the enzyme contains a second site to which an upstream region of the primer remains anchored as the active site moves along the template (Fig. 4). In particular, the second site was proposed to be required as an anchor for the primer during translocation, at which stage the Watson-Crick hydrogen bonds between the template and the primer are thought to be completely disrupted.
Detailed Analysis of Primer Extension Performed with TER Mutants
More recently, studies of primer extension by the Tetrahymena telomerase were carried out with various telomerase mutants, including mutants in the RNA subunit of the enzyme. The latter type of mutants were synthesized in cells transfected with plasmids encoding mutant RNAs, or were reconstituted in vitro, as described in the previous section. Studies performed with the various RNA mutants led to a precise definition of the RNA template region.
Nucleotides C49-C43 were found to act as templating residues, that is, to be copied into the complementary DNA nucleotides (see Fig. 4). This conclusion was based on data obtained in assays of mutants in which individual RNA residues were substituted with other residues.28–30 For example, unlike the wild-type enzyme, a mutant designated 43A, in which the residue C43 had been replaced with A, incorporated a T residue at the corresponding position in the extended primer.28 Similarly, a mutant designated 49G, in which the residue C49 had been replaced with G, incorporated a C residue at the corresponding position in the primer.29
The RNA template region was also found to include the two A residues, A50 and A51. These residues are not employed as templating nucleotides, as indicated by the observation that primers aligned upstream of these positions could not be extended.29 However, they play essential roles in the primer extension reaction. The role of the RNA nucleotide no. 50 was indicated by assays performed with a mutant designated 50G, in which the residue 50A had been replaced with G. This mutant enzyme was found to be very inefficient in extending primers ending with the sequence TGGG-3'. However, the efficiency of the extension reaction was greatly enhanced in a similar reaction performed with the 50G mutant and a primer ending with the sequence CGGG-3', in which the C residue rather than T faced the 50G residue in the template. Thus, formation of a canonical Watson-Crick base pair between the primer and the template at the nucleotide position no. 50 is essential for the elongation process.29 Similar results were obtained in assays that were performed with an enzyme in which the RNA residue A51 was mutated. However, this mutation was less inhibitory to elongation of oligonucleotides that did not form canonical Watson-Crick base pairs with the RNA residue A51. Thus, the RNA residues A50 and A51 are required for proper alignment of the primer along the template, even though they are not used as templating nucleotides.
The studies performed with telomerase RNA mutants also indicated that, besides templating, the interactions between the RNA nucleotides in the template region and DNA primer residues also affect other aspects of the elongation process. For example, the 43A mutation discussed above caused premature product dissociation at position 45 along the template even before this nucleotide was reached by the active site of the enzyme. Another mutation, designated 48U, caused both product dissociation at position 45 and misincorporation of A residues at positions 47 and 46.29 Thus, this substitution mutation at the template position 48 caused a greatly diminished fidelity of incorporation at other positions along the template. These results implied that specific bases within the template region play essential roles in active site functions during primer elongation.
In addition to the single nucleotide substitution mutations discussed above, telomerase RNA mutants, in which the entire template region was substituted with non-telomeric sequences, were also generated. Two such mutants designated AUN and U9, contained the sequences: 3'-aaAAUAUAUAUuu-5' and 3'-aaUUUUUUUUUuu-5', instead of the wild-type template sequence 3'-aaAACCCCAACuu-5' (The capital letters designate the templating nucleotides; see Fig. 4). These mutant enzymes were found to be catalytically active and to incorporate the proper complementary nucleotides, i.e., dATP and dTTP, during primer extension reactions.52 However, unlike the wild-type enzyme, after completing the first round of elongation these mutants were unable to reposition the 3' termini of the primers at the proper site opposite the 3' end of the template region and therefore were not processive. Moreover, the AUN mutation caused telomere shortening in vivo that apparently led to impaired cell growth.
Primer extension experiments performed with the reconstituted Tetrahymena telomerase provided information on the roles that regions of TER outside the template region played in the primer extension process. The sequence 5'-CUGUCA-3', located two nucleotides upstream of the 5' end of the template region, was found to be conserved in all ciliates.25–27 In most ciliates, including Tetrahymena, the 5' end of this sequence is found in stem-loop II (see Fig. 1). It was observed that some mutations introduced into this sequence caused defects in the binding of the RNA to TERT32 (see above section). Other mutations affected certain aspects of the elongation process. For example, substitution of the three nucleotides UCA at the 3'-end of the conserved sequence by AGU caused a substantial reduction in enzyme activity, and only a moderate inhibition in the binding of TERT to TER.32 This mutant enzyme also copied the RNA past the normal 5' template boundary at nucleotide C43 and was less processive.32,53 These data and assays of other mutants in the conserved sequence suggested that this sequence is involved in the definition of the 5' boundary of the template and in the signal that leads to the translocation step.32,53
As noted in a previous section, RNA footprinting of in vitro reconstituted enzyme complexes revealed the involvement of stem-loop IV in the folding of the pseudoknot during the assembly of the telomerase RNP.33 However, mutations in loop IV also caused a severe inhibition of telomerase activity that could not be accounted for entirely by the defect in the assembly process. Hence, it has been suggested that loop IV, which is distant from the template region in the RNA chain (Fig. 1), also directly affects the function of the active site of the telomerase.33
Analysis of Primer Extension Performed with TERT Mutants
As already mentioned above, it now appears that TERT consists of at least two separate domains: a reverse transcriptase domain and an RNA binding domain, which strongly binds regions of TER other than the template region. The RT domain is probably formed from the sequences in and around the RT motifs (Fig. 2). This domain contains the active site of the enzyme, of which the three conserved aspartates in motifs A and C are important constituents.17,22,40
Additional features of the RT domain and of the RNA binding domain of TERT have been recently studied by primer extension assays performed with Tetrahymena telomerase TERT mutants.20,36,37,54 These mutant enzymes were assembled in vitro in rabbit reticulocyte lysates, as described above. It was found that mutations in motifs 1, 2 of TERT caused a decrease in processivity of the enzyme during the synthesis of a single repeat and also inhibited the initiation of another repeat. Mutations in motifs A and D inhibited the extension of primers aligned next to the 5' end of the template. Certain mutations in motif A also caused the enzyme to be less selective for dNTPs vs. rNTPs. It should be noted that a similar decrease in the discrimination between deoxyribonucleotides and ribonucleotides was found to be caused by mutations introduced into corresponding amino acids in motif A of viral reverse transcriptases.54 It was concluded from these studies that several features of the structure-function relationships of the RT domain in TERT are similar to those of the viral RTs. However, the RT domain of TERT has also evolved to carry out the specific telomerase functions, such as the definition of the 5' and 3' boundaries of the template.
Some functions of the RT domain of the Tetrahymena TERT are also affected by interactions with the other domain (or domains) of TERT; for mutations in the CP2 motif, were found to partially abolish the 5' template boundary. Furthermore, non-conserved regions near the amino and the carboxyl termini of TERT are also needed for the reverse transcriptase activity of the enzyme.37 As pointed out in the previous section, nucleotide residues in the RNA template region also play a role in active site functions. Therefore, it appears that these nucleotides interact with specific amino acids residues of the TERT reverse transcriptase domain to ensure proper alignment and extension of the primers by the enzyme.
The Special Role of dGTP in Primer Extension by Telomerase
Kinetic studies performed with the Tetrahymena telomerase revealed that the Km for dGTP incorporation into the primers was about 10-fold lower than the Km for dTTP incorporation.55,56 Moreover, in addition to serving as a precursor for the primer extension reaction, dGTP was found to specifically enhance the processivity of the enzyme. This additional function of dGTP has been extensively characterized in studies performed with the reconstituted telomerase.57 It is specific for the G base and the sugar deoxyribose, since other dNTPs, or GTP, did not stimulate the processivity of the enzyme. The processivity was also enhanced by dGMP, but not by guanosine. Thus, triphosphate hydrolysis was not required for this function, but the presence of a phosphate group was essential. Further experiments indicated that the processivity-stimulatory dGTP molecule binds at the active site for the elongation of the primer and not at a separate site.54 It should also be noted that the reconstituted core enzyme used for these studies was considerably less processive than native telomerase. Hence, the native telomerase appears to possess an additional mechanism for processive elongation of the primers.22 dGTP was also found to stimulate the processivity of the telomerase of another ciliate, Euplotes aediculatus. However, in this ciliate the dGTP that enhances the processivity might bind a site in the enzyme which is distinct from the active site for polymerization.58
Finally, it is noteworthy that in vivo the Tetrahymena telomerase may not extend telomeres by a processive mechanism of the type observed in vitro. For Tetrahymena cells containing both a wild-type telomerase and a mutant in the templating nucleotides, newly synthesized wild-type and mutant telomeric repeats were found to be interspersed.19
Extension of Primers That Contain Non-Telomeric Repeats and Implications for de Novo Telomere Synthesis
Early studies of the Tetrahymena telomerase have indicated that single-stranded DNA primers lacking telomeric repeats (non-telomeric primers) are very poor substrates for the enzyme.59,60 Primers containing telomeric repeats at the 3' termini and non-telomeric repeats at the 5' termini were, however, efficiently extended by the telomerase.60 Evidently, these primers could properly align with the RNA template region, like the telomeric primers. Moreover, primers containing telomeric repeats at the 5' termini and non-telomeric sequences at the 3' termini were also extended by the enzyme at a rather high efficiency, even though the 3' ends of these primers could not form canonical Watson-Crick hydrogen bonds with the nucleotides in the RNA template region.60 One possible explanation for these data was that the G-rich telomeric repeats at upstream positions in the primers have a higher affinity for the hypothetical second site of the enzyme than non-telomeric sequences (see Fig. 4). The interaction with the second site was proposed to increase the probability of extension of the non-telomeric 3'-termini in these primers.13,60 This interpretation has been recently tested by primer extension assays carried out with the telomerase RNA mutants described above, in which the entire RNA template region was substituted with a non-telomeric template sequence.52 The primers used for these assays contained a 3'-terminal sequence complementary to the mutated RNA template. Upstream of this sequence the primers contained various arrangements of Tetrahymena telomeric repeats, (TTGGGG)n, and non-telomeric sequences. The data obtained in these experiments indicated that primers containing G-rich telomeric repeats located between nucleotides 12–17 upstream of the 3' termini were more efficiently extended than primers containing non-telomeric DNA at these positions. These data appeared consistent with preferential telomerase second site binding of authentic telomeric repeats at upstream positions in the primers. However, it should be pointed out that the mutant enzymes were not processive and that, therefore, these conclusions may not apply to the processive mode of primer extension of the wild-type enzyme.
The question whether non-telomeric primers can be extended by the telomerase was reexamined in a series of studies, in which the conditions used for the reactions were carefully monitored.61 These studies revealed that the Tetrahymena telomerase can extend primers that do not contain any telomeric repeats at nanomolar concentrations, albeit at lower efficiencies than the efficiencies with which primers containing telomeric repeats were extended.61 In particular, the telomerase was capable of extending non-telomeric primers containing sequences that are apparently extended in vivo by the telomerase during chromosome healing events. The elongation of the non-telomeric primers was found to be especially sensitive to concentrations of mono- and divalent ions in the reaction mixtures, and this could explain the apparent discrepancy with some of the earlier results, which indicated that non-telomeric primers could not be extended. Tetrahymena telomerase isolated from mated cells has specificities for non-telomeric and telomeric primers that are indistinguishable from enzyme isolated from vegetative cells. In contrast, telomerase purified from developing macronuclei in cells from another ciliate, Euplotes crassus, was found to contain a factor needed for elongation of non-telomeric 3'-ends.62 This factor was absent from telomerase purified from vegetative E. crassus cells. It should be noted that both the Tetrahymena and the Euplotes telomerases initiate the extension of non-telomeric primers with the sequence GGGGT-3'.
The length dependence of the efficiency with which non-telomeric primers are extended by the Tetrahymena telomerase was found to be substantially different from the length dependence observed for telomeric primers. All single-stranded telomeric primers that were longer than 10–12 nucleotide residues were found to be efficiently extended by the enzyme.55 In contrast, non-telomeric primers containing less than 20 nucleotide residues were not efficiently elongated.61 Furthermore, the efficiency of elongation increased substantially as the number of residues in the non-telomeric primers increased up to about 30 nucleotides. As previously discussed, the telomerase is capable of elongating single-stranded 3'-overhangs of double-stranded DNA molecules. It is, however, incapable of extending double-stranded DNA that does not end with 3'-overhangs.61 The minimal length of an overhang consisting of telomeric repeats that could be extended by the Tetrahymena telomerase was 13 nucleotides.63 For non-telomeric 3' overhangs, the minimal length allowing efficient extension was around 20 nucleotides.61 It should also be noted that telomeric 3'-extensions of 4–6 nucleotides sufficed for efficient elongation by another ciliate telomerase, that of Euplotes aediculatus.64 These data indicated that the interactions between the telomerase and non-telomeric primers, which occur during the initial stages of the elongation of such primers, differ from the interactions that occur between the enzyme and telomeric primers. Further discussion of these differences is presented below.
The Telomerase Nucleolytic Activity
The Tetrahymena telomerase has an inherent nucleolytic activity. This activity was discovered in studies of extension of primers ending with the sequence GGGGTT-3', which aligned with the telomerase RNA near the 5'-end of the template region65 (see Fig. 4). Extension of these primers by the telomerase in the presence of 32P-dGTP and ddTTP, gave rise to relatively abundant elongation products containing 5 newly added nucleotide residues, of which the product GGGGTTg-3' was the most abundant. A study of the time course of accumulation of this product showed that after reaching a maximal level, its concentration decreased. Additional experiments revealed that the decrease resulted from removal of the 3' G residue. Further studies confirmed that this removal was due to an intrinsic 3' nucleolytic activity of the telomerase and that nucleotide cleavage occurred primarily in primers that were aligned near the 5' end of the RNA template region.65 Both correct complementary nucleotides and incorrect non-complementary residues were found to be removed by the telomerase nucleolytic activity. Thus, the telomerase 3'-nuclease does not appear to be primarily utilized for proofreading, unlike the 3'-exonuclease activities of various DNA polymerases.53,65,66
Another ciliate telomerase, that of Euplotes crassus, was also found to have a 3' nucleolytic activity.67 In this study, the telomerase was found to have endonuclease activity, rather than a 3'-exonuclease. A further characterization of the Euplotes telomerase nuclease activity revealed that it was capable of removing fragments consisting of several nucleotides from the 3' termini of primers containing a non-telomeric sequence at the 3' terminus and a telomeric sequence further upstream. It has been proposed that this telomerase possesses a distinct cleavage active site, which is separated from the polymerase site. The primers align with the template at the cleavage site, such that the 3' non-telomeric sequence is removed. Subsequently, the telomeric repeat which becomes the 3' sequence, is realigned along the RNA template region at the polymerase site, and the primers are extended by new telomeric sequences.67 The proposed nucleolytic activity of telomerase appears to be analogous to the nucleolytic activity of RNA polymerases.68,69 Both the telomerase and the RNA polymerase activities might act to remove sequences that interfere with extension of the primers (or the RNA transcripts). Another possible function of the telomerase nuclease activity might be to prevent the enzyme from transcribing RNA nucleotides outside the template region.67
UV Crosslinking Analysis of Interactions Between the Telomerase and the DNA Primers
Crosslinking assays of complexes formed between the telomerase and DNA primers were carried out with the photoreactive nucleotide analogue, 5-[N-(p-azidobenzoyl)-3-aminoallyl]-deoxyuridine triphosphate] (N3RdUTP).42,70 This compound was found to be a substrate for primer extension by the telomerase.42 UV crosslinking of elongation complexes containing primers that incorporated N3RdU, followed by SDS-polyacrylamide gel electrophoresis of the irradiated complexes, revealed a 100 kD protein that became crosslinked to the DNA. This protein has also been identified by crosslinking the Tetrahymena telomerase with iodouracil-substituted DNA primers.42 More recent UV crosslinking assays of the Tetrahymena telomerase elongation complexes performed with the reagent N3RdUTP revealed a 145-kD band in the gel. This band presumably contained the 133-kD TERT crosslinked to the DNA primer (Manor, H., unpublished data).
UV crosslinking was also employed for studies of interactions between DNA primers and the telomerase of the ciliate Euplotes aediculatus.71 These studies were performed with single-stranded DNA molecules and with double-stranded DNA molecules containing 3'-overhangs, in which various nucleotide residues had been substituted with photoreactive 5-iododeoxypyrimidines. The primers were found to crosslink to a 130-kD protein, probably the Euplotes TERT, and to specific residues in the RNA subunit (U51-U52). Detailed mapping of the interactions further revealed that in the primers the crosslinks occurred with nucleotides nos. 20 to 22 upstream of the 3'-end. It has been proposed that these crosslinks represented interactions of the primers with the anchor site of the enzyme.71 Similar UV crosslinking assays have been recently performed with FLAG-tagged Tetrahymena TERT that had been synthesized in rabbit reticulocyte lysates and attached to beads through the tag. DNA primers substituted with 5-iododeoxyuridine were found to crosslink not only to the reconstituted enzyme including both TERT and TER, but also to TERT alone.33
Interference Footprinting Analysis of Telomerase Elongation Complexes
An interference footprinting analysis of Tetrahymena telomerase elongation complexes has provided information on functional interactions between single-stranded telomeric DNA primers and the telomerase RNP.72 In this study, the primers were first modified with base-specific chemical reagents. The modified primers were extended with telomerase by a single 32P-labeled dNMP, thereby creating a population of end-labeled fragments. Base modifications that interfered with the primer extension reactions were mapped by footprinting. Major functional interactions were detected between the enzyme and the six or seven 3'-terminal residues of the primers. Only weak interactions occurred with nucleotide residues located further upstream in these primers. Moreover, the strengths of these interactions were not substantially affected by the presence or absence of telomeric sequences at upstream positions in the primers. Similar data were obtained with Tetrahymena core telomerase that had been reconstituted by coexpression of TERT and TER in rabbit reticulocyte lysates (Baran N, Haviv Y and Manor H, in preparation). Hence, the interactions that were previously detected with the native telomerase occurred primarily with the core region of the enzyme.
Interference footprinting assays were also performed with core telomerase assembled from wild-type TERT and RNA substitution mutants, as described above. The use of the RNA mutants allowed footprinting of primers that were aligned at four different positions along the template region. These studies indicated that as the primers are elongated, the region in the telomerase RNP that interacts with the 3'-terminal nucleotides moves relative to the template region in the RNA, along with the active site of the enzyme (Baran N, Haviv Y and Manor H, in preparation).
Updated Model for Primer Extension by Telomerase
In view of the data discussed in this review, we propose the model illustrated in Figure 5 for extension of single-stranded telomeric DNA primers by telomerase. This model assumes that TERT consists of at least two separate domains, the reverse transcriptase domain and another domain designated here as the second domain, which strongly binds regions in TER other than the template region (see refs. 36,37,41). The 3'-end of a telomeric primer aligns with the template region in TER and the active site of the enzyme, as indicated (stage I). As the elongation proceeds, the active site of the enzyme must be displaced relative to the template region. We propose that it is the reverse transcriptase domain of TERT, which includes the active site, that moves relative to the template and the second domain of TERT (stage II). We also propose that at each stage in the elongation process, up to six or seven 3'-terminal nucleotide residues in the primers are associated with the reverse transcriptase domain and with the template. When the elongation proceeds as far as the 5' terminus of the template region (stage III), the 3'-end of the primer is detached from the RNA and is translocated to the 3' terminus of the template. The active configuration of the enzyme-primer complex is reestablished at this stage (stage IV) and synthesis of the next telomeric repeat is initiated. It is not clear at the present time what type of interactions occur between the enzyme and the primer during the translocation. It is possible that significant interactions occur at this stage between nucleotides located upstream of the seventh residue in the primer and the second domain of TERT, which serves as the anchor. It is also possible that even though the 3'-end of the primer is detached from the template, it is not detached from the reverse transcription domain of TERT and both coordinately move to the 3'-end of the template region.
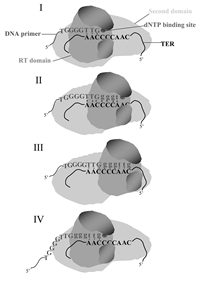
Figure 5
Updated model for extension of telomeric primers by the Tetrahymena telomerase. Details are presented in the text.
The model discussed here is compatible with the genetic and the footprinting data and with the length dependency of the efficiency of extension of telomeric primers, as discussed in this review. It should be noted that separation of the catalytic center from a domain that strongly binds the RNA outside the template region, and the relative movement of these regions during primer elongation, were features of previous models proposed for telomerase.36,73
The mechanism of extension of non-telomeric primers is less clear and is not presented here as a drawing. Apparently, during the initiation of such primers, which cannot interact with the template, different interactions with the enzyme align these primers next to the template, such that the sequence GGGGTT is synthesized first. Such alignment apparently requires that the primers be longer than telomeric primers.15,61 However, after the first round of synthesis, the 3'-ends of these primers are realigned with the enzyme, such that further extension proceeds as illustrated in Figure 5.
Clearly, additional research is required to examine the various features of the model proposed in Figure 5. In particular, it would be necessary to obtain a more detailed information on the interactions of the DNA primers with the telomerase during the various stages of the primer extension process.
Acknowledgments
The research performed in the authors' laboratory was supported by The Israel Science Foundation (grant No. 373/99).
References
- 1.
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–58. [PubMed: 11092831]
- 2.
- Collins K. Mammalian telomeres and telomerase. Curr Opin Cell Biol. 2000;12:378–383. [PubMed: 10801465]
- 3.
- Evans SK, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J Cell Sci. 2000;113:3357–64. [PubMed: 10984427]
- 4.
- Shore D. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr Opin Genet Dev. 2001;11:189–98. [PubMed: 11250143]
- 5.
- Li B, Oestreich S, de Lange T. Identification of human Rap1:implications for telomere evolution. Cell. 2000;101:471–83. [PubMed: 10850490]
- 6.
- Watson JD. Origin of concatameric T7 DNA. Nature New Biol. 1972;239:197–201. [PubMed: 4507727]
- 7.
- Olovnikov AM. A theory of marginotomy. J Theor Biol. 1973;41:181–90. [PubMed: 4754905]
- 8.
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. [PubMed: 3907856]
- 9.
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–7. [PubMed: 2463488]
- 10.
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–60. [PubMed: 2342578]
- 11.
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–52. [PubMed: 9454332]
- 12.
- Blackburn EH. Developmentally Programmed Healing of Chromosomes In: Telomeres, E.H. Blackburn and C.W. Greider (ed).Cold Spring Harbor Laboratory Press,1995. p.193–218.
- 13.
- Morin GB. Recognition of a chromosome truncation site associated with alpha- thalassaemia by human telomerase. Nature. 1991;353:454–6. [PubMed: 1896089]
- 14.
- O'Reilly M, Teichmann SA, Rhodes D. Telomerases. Curr Opin Struct Biol. 1999;9:56–65. [PubMed: 10047586]
- 15.
- Collins K. Ciliate telomerase biochemistry. Annu Rev Biochem. 1999;68:187–218. [PubMed: 10872448]
- 16.
- Blackburn EH. The end of the (DNA) line. Nat Struct Biol. 2000;7:847–850. [PubMed: 11017190]
- 17.
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–7. [PubMed: 9110970]
- 18.
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–95. [PubMed: 9288757]
- 19.
- Yu GL, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–32. [PubMed: 1689810]
- 20.
- Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J Biol Chem. 2000;275:24199–207. [PubMed: 10807925]
- 21.
- Weinrich SL, Pruzan R, Ma LB, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. [PubMed: 9398860]
- 22.
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci U S A. 1998;95:8485–90. [PMC free article: PMC21102] [PubMed: 9671704]
- 23.
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, Wright WE, White MA. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–26. [PMC free article: PMC316592] [PubMed: 10197982]
- 24.
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J. et al. The RNA component of human telomerase. Science. 1995;269:1236–41. [PubMed: 7544491]
- 25.
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–53. [PubMed: 1840508]
- 26.
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–98. [PubMed: 7958872]
- 27.
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–7. [PMC free article: PMC306816] [PubMed: 7739888]
- 28.
- Gilley D, Lee MS, Blackburn EH. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–26. [PubMed: 7557376]
- 29.
- Gilley D, Blackburn EH. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. [PMC free article: PMC230979] [PubMed: 8524330]
- 30.
- Autexier C, Greider CW. Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev. 1994;8:563–75. [PubMed: 7523243]
- 31.
- Gilley D, Blackburn EH. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci U S A. 1999;96:6621–5. [PMC free article: PMC21964] [PubMed: 10359761]
- 32.
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 1999;13:1116–25. [PMC free article: PMC316941] [PubMed: 10323863]
- 33.
- Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–16. [PubMed: 11401544]
- 34.
- Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–14. [PubMed: 10721988]
- 35.
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–5. [PubMed: 10591218]
- 36.
- Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–9. [PubMed: 10983995]
- 37.
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. [PMC free article: PMC99554] [PubMed: 11158287]
- 38.
- Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–90. [PubMed: 9506510]
- 39.
- Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol. 1998;8:54–63. [PubMed: 9519297]
- 40.
- Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci U S A. 1998;95:8479–84. [PMC free article: PMC21101] [PubMed: 9671703]
- 41.
- Bachand F, Autexier C. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol Cell Biol. 2001;21:1888–97. [PMC free article: PMC86762] [PubMed: 11238925]
- 42.
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–86. [PubMed: 7774009]
- 43.
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–84. [PubMed: 9118230]
- 44.
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science. 1997;275:973–7. [PubMed: 9020079]
- 45.
- Liu Y, Snow BE, Hande MP, Baerlocher G, Kickhoefer VA, Yeung D, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, Harrington L. Telomerase-Associated Protein TEP1 Is Not Essential for Telomerase Activity or Telomere Length Maintenance In Vivo. Mol Cell Biol. 2000;20:8178–84. [PMC free article: PMC86427] [PubMed: 11027287]
- 46.
- Miller MC, Collins K. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol Cell. 2000;6:827–37. [PubMed: 11090621]
- 47.
- Forsythe HL, Jarvis JL, Turner JW, Elmore LW, Holt SE. Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J Biol Chem. 2001;276:15571–574. [PubMed: 11274138]
- 48.
- Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–800. [PMC free article: PMC316652] [PubMed: 9353249]
- 49.
- Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–34. [PMC free article: PMC125520] [PubMed: 11432839]
- 50.
- Greider CW. Telomerase biochemistry and regulation In:Telomeres, E.H. Blackburn and C.W. Greider (ed). Cold Spring Harbor Laboratory Press, 199535–68.
- 51.
- Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–80. [PMC free article: PMC361337] [PubMed: 1875940]
- 52.
- Ware TL, Wang H, Blackburn EH. Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J. 2000;19:3119–31. [PMC free article: PMC203363] [PubMed: 10856255]
- 53.
- Autexier C, Greider CW. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–39. [PubMed: 7557377]
- 54.
- Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–22. [PMC free article: PMC302041] [PubMed: 10944124]
- 55.
- Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–99. [PMC free article: PMC364717] [PubMed: 8413255]
- 56.
- Collins K, Greider CW. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J. 1995;14:5422–32. [PMC free article: PMC394651] [PubMed: 7489731]
- 57.
- Hardy CD, Schultz CS, Collins K. Requirements for the dGTP-dependent Repeat Addition Processivity of Recombinant Tetrahymena Telomerase. J Biol Chem. 2001;276:4863–71. [PubMed: 11096070]
- 58.
- Hammond PW, Cech TR. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 1997;25:3698–704. [PMC free article: PMC146957] [PubMed: 9278493]
- 59.
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–98. [PubMed: 3319189]
- 60.
- Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–4. [PubMed: 1896088]
- 61.
- Wang H, Blackburn EH. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–79. [PMC free article: PMC1169687] [PubMed: 9049315]
- 62.
- Bednenko J, Melek M, Greene EC, Shippen DE. Developmentally regulated initiation of DNA synthesis by telomerase: Evidence for factor-assisted de novo telomere formation. EMBO J. 1997;16:2507–18. [PMC free article: PMC1169850] [PubMed: 9171363]
- 63.
- Lee MS, Gallagher RC, Bradley J, Blackburn EH. In vivo and in vitro studies of telomeres and telomerase. Cold Spring Harb Symp Quant Biol. 1993;58:707–18. [PubMed: 7956088]
- 64.
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3' overhang. Proc Natl Acad Sci USA. 1996;93:10712–17. [PMC free article: PMC38220] [PubMed: 8855245]
- 65.
- Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–76. [PubMed: 8330740]
- 66.
- Kornberg A, Baker TA. DNA Replication, 2 edn. W. H. Freeman and Company, 1992.
- 67.
- Melek M, Greene EC, Shippen DE. Processing of nontelomeric 3' ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–45. [PMC free article: PMC231338] [PubMed: 8668159]
- 68.
- Surratt CK, Milan SC, Chamberlin MJ. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci U S A. 1991;88:7983–7. [PMC free article: PMC52429] [PubMed: 1716768]
- 69.
- Reines D, Ghanouni P, Li QQ, Mote J Jr. The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992;267:15516–15522. [PMC free article: PMC3371615] [PubMed: 1379232]
- 70.
- Harrington L, Hull C, Crittenden J, Greider C. Gel shift and UV cross-linking analysis of Tetrahymena telomerase. J Biol Chem. 1995;270:8893–901. [PubMed: 7721797]
- 71.
- Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. [PMC free article: PMC231754] [PubMed: 8972210]
- 72.
- Benjamin S, Baran N, Manor H. Interference footprinting analysis of telomerase elongation complexes. Mol Cell Biol. 2000;20:4224–37. [PMC free article: PMC85791] [PubMed: 10825187]
- 73.
- Wang H, Gilley D, Blackburn EH. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–60. [PMC free article: PMC1170463] [PubMed: 9463392]
- DNA Primer Extension by Telomerase - Madame Curie Bioscience DatabaseDNA Primer Extension by Telomerase - Madame Curie Bioscience Database
- JGI_CAAK5209.rev NIH_XGC_tropBrn3 Xenopus tropicalis cDNA clone IMAGE:7655698 3'...JGI_CAAK5209.rev NIH_XGC_tropBrn3 Xenopus tropicalis cDNA clone IMAGE:7655698 3', mRNA sequencegi|58404174|gnl|dbEST|27518750|gb|C 41.1|Nucleotide
Your browsing activity is empty.
Activity recording is turned off.
See more...