NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The conformational flexibility of RNA is the basis for its functional versatility. RNA molecules can fold into functionally diverse structures, providing the grounds for great regulatory potential. RNA function can be turned on or off in very short time scales merely through, for example, the binding of ligands. Conformational switching of RNA molecules can be induced by diverse signals, ranging from large molecules like proteins and RNAs to small molecules like coenzymes, amino acids, or nucleotides to temperature and the translation speed of the ribosome. In this review, we discuss switches for RNA folding and function induced by naturally occurring RNA-protein interactions.
The conformational flexibility of RNA is also the basis for the RNA folding problem. The numerous structures that an RNA molecule encounters on its folding pathway represent kinetic traps, which can slow down the folding process significantly. In nature, a large number of proteins with different modes of action help RNA molecules to switch conformations. These proteins can stabilize specific structures or nonspecifically resolve low stability conformers. The latter activity has been called RNA chaperone activity, a function that has been attributed to a large and ever-growing number of proteins. Specialized RNA helicases hydrolyze ATP to resolve stable structures. Noncoding RNAs, which induce functional switches by base pairing, have difficulties identifying their targets due to the generally low accessibility of supposedly unpaired bases. The E. coli protein Hfq helps noncoding RNAs to solve this problem. There are many ways by which proteins participate in and regulate RNA structure and as a consequence its function. The diversity of these mechanisms is just beginning to be understood and will be discussed in this review.
Introduction
RNA molecules are linear polyanionic chains that must fold into defined three-dimensional structures to accomplish tasks as diverse as catalysis, ligand binding, and protein recognition. The folding pathway of RNA molecules is hierarchical, starting with the formation of short double-stranded helices via Watson-Crick (WC) type base pairs, which are part of the secondary structure. These short secondary structure elements then organize themselves in space to form very diverse tertiary structures.1 Apart from the canonical AU and GC pairs, RNA supports GU pairs, so the number of possible pairing combinations is very high and the alternative conformations are often comparably stable. This structural versatility is a challenge for an RNA molecule, because it becomes more difficult to reach its native folding state.2 On the other hand, this structural promiscuity opens the way to an important attribute: the possibility for one RNA molecule to exist in different conformational states achieving different functions. An extreme example of an RNA sequence that folds into two functionally completely different molecules is a designed variant of an in vitro selected RNA ligase that can also fold into a hepatitis delta virus ribozyme. This RNA sequence has two identities, that of an artificial RNA ligase and that of an HDV ribozyme.3
The notion that RNAs have flexible conformations arose with the discovery of attenuation at the tryptophan operon by the pioneering work of Charles Yanofski.4 Several decades later, the phenomenon of structural variation of messenger RNA induced by small ligands and leading to different functions was discovered. These naturally occurring structural rearrangements induced by small molecules have been termed “riboswitches” (see Chapter 9). In the past few years, an unexpectedly large number of riboswitches have been discovered. Ron Breaker's laboratory in particular published a series of papers describing RNA conformational switches induced by small metabolites.5-7 Binding of these small molecules to the 5'-untranslated region of an mRNA leads to conformational changes resulting in translation inhibition, transcription attenuation, or RNA cleavage. In general, the signals that can induce a switch in RNA conformation and function are diverse, ranging from large molecules like proteins or tRNAs, small molecules like cobalamin, FMN, guanosine or lysine to temperature, the speed of transcription or of the translating ribosome.8-10 Small, noncoding RNAs have also been reported to induce switches in RNA conformation to regulate mRNA stability and translation.11
By summarizing and comparing the latest experiments, this review discusses how proteins can induce conformational switches to regulate the function of RNA molecules. We distinguish between different classes of proteins according to their mode of action. A protein can alter the structure of an RNA molecule in several ways. If the RNA is structurally unstable, leading to a mixed population of conformers, binding of a specific protein can stabilize one conformation, thus favoring it over the others. Tight binding of a protein to one part of the RNA might not be enough to unfold stable alternative conformations, requiring the unwinding activity of RNA helicases. In such a case, exemplified by the Neurospora crassa tyrosyl tRNA synthetase that recognizes group I intron RNAs, the specific RNA-binding protein recruits a specific RNA helicase, which consumes ATP to unfold the nonnative structure.12 Another type of protein activity is attributed to Hfq, which binds RNA molecules less specifically and induces the unfolding of neighboring domains, making these accessible to additional binding partners. Sm and Sm-like proteins, which are required for the conformational flexibility of large functional RNAs, may share some properties with Hfq. Finally, we discuss a large family of proteins, such as the nucleocapsid peptide NCp7 from HIV and the E. coli StpA protein, that have RNA chaperone activity.13,14 These proteins are diverse in function but have the common property that they resolve RNA structures in a rather nonspecific way.
Proteins That Bind RNA with High Affinity and Stabilize Specific Structures
In nature, most RNA-based reactions take place in conjunction with proteins. Many RNAs are associated with a specific protein factor that binds tightly to a defined structure, stabilizing it and reducing the population of alternative conformations that act as folding traps (fig. 1). Studying protein-dependent RNA folding is tedious if the folded state of the RNA has to be determined for each step. Therefore, the use of catalytic RNAs has proven to be especially suitable, because the native state correlates with function. Self-splicing introns that depend on protein cofactors are good systems for addressing the question of how a protein binds to a specific RNA target and how it aids in the folding process. For example, the group I intron-specific splicing factor Cyt-18 is a dimeric tRNA synthetase, which stabilizes the group I intron core by binding the P4-P6 domain.15 In contrast, the maturase I-AniI encoded by the Aspergillus nidulans mitochondrion binds rapidly to the COB preRNA, forming a specific yet labile encounter complex that must be resolved to form a native functional complex in a rate-limiting step. The maturase preassociates with the unfolded RNA and probably facilitates correct folding by resolving misfolded RNAs or by reducing the number of possible conformations; after the intron is correctly folded, the protein locks the RNA in its native state by specific binding.16 A third example of a specific interaction between a protein and its target group I intron RNA is the yeast CBP2 protein, which facilitates splicing of the mitochondrial bI5 intron. The bI5 intron has been shown to form a collapsed state prior to folding into the native state in the presence of 7 mM Mg2+, whereas it forms an expanded structure at low Mg2+ concentrations.17 The CBP2 protein binds to the intron RNA, which is in its roughly globular native-like collapsed state, and converts it into the native state without mediating global conformational rearrangements, instead promoting only the final few angstroms of RNA folding.18 Specific RNA-binding proteins have in common that they form tight complexes with their target RNAs, but how these proteins bind and how they guide the RNA to their native states can vary substantially.
RNA Helicases
DexD/H-box proteins are putative RNA helicases that unwind double stranded RNA (dsRNA) in an energy-dependent fashion using a nucleoside triphosphate, preferentially ATP.19-22 Two groups of proteins are RNA helicases: the DEAD-box and the DexH-box proteins. Both are named according to their conserved amino acid sequence motifs.23,24 From the large number of DexD/H proteins, only a few have been proven to possess RNA helicase activity.25 Putative RNA helicases are present in almost all organisms ranging from bacteria and viruses to humans and are involved in a large variety of cellular processes, including nuclear transcription, premRNA splicing, ribosome biogenesis, nucleocytoplasmic transport, translation, RNA decay, and organellar gene expression.26 The best-studied DEAD-box protein is the eukaryotic initiation factor eIF4A.27,28 Some of these proteins may even be involved in disrupting or rearranging RNA-protein interactions.21,25,29 In contrast to DNA helicases, the RNA counterparts are thought to modulate formation of only short RNA duplexes in a one-step reaction.
There are similarities in the tertiary structures of DNA and RNA helicases.30 In vitro, RNA helicases show only limited substrate specificity, whereas in vivo, specificity is necessary in order to prevent random opening of RNA structures and RNA-protein interactions. Therefore, RNA helicases need a specificity factor to identify their place of action (fig. 2). From the very large number of RNA helicases, we focus here on two interesting exemplary studies: the active disruption of an RNA-protein interaction by the viral DexH/D RNA helicase NPH-II, and the DEAD-box protein Cyt-19 from Neurospora crassa that uses Cyt-18 as specificity factor for finding its substrate.12,25
NPH-II
The first in vitro demonstration of the active disruption of an RNA-protein interaction was shown with the vaccinia virus NPH-II DexH/D helicase.25 The disruption of an RNA-protein interaction has been named RNPase or RNP displacement activity. In this report, the well-characterized U1A protein and its natural target binding site, the 3'-UTR of the U1A mRNA, were used.31 U1A binds to this target as a dimer, interacting with two asymmetric loops. In order to fulfill its function as an RNA helicase substrate, the RNA was altered by removing a hairpin loop and extending the flanking helical regions. A 3'-overhang was also included to provide a platform for NPH-II binding. This altered substrate still binds U1A very tightly. Using this system, a basic kinetic mechanism of helicase action was established. The dissociation rate for U1A was increased by several orders of magnitude when NPH-II and ATP were present. Although NPH-II appears to “stumble” when it encounters the U1A protein, it maintains its processivity.21 Thus, NPH-II is able to displace U1A and continue unwinding the RNA substrate, without falling off during the course of reaction.
Cyt-19
Cyt-19 is a DEAD-box protein isolated from Neurospora crassa using a cold-sensitive group I intron splicing-deficient mutant.12,32 As expected for a DEAD-box protein, Cyt-19 is an RNA-dependent ATPase. The purified protein does not specifically bind to the group I intron RNA but requires the group I intron specific splicing factor Cyt-18 as specificity factor. Thus, this RNA helicase functions only in concert with Cyt-18 to promote group I intron splicing. The addition of Cyt-19 and ATP increases both the rate of splicing and the total yield of spliced RNA, suggesting that Cyt-19 resolves misfolded RNAs. Using the well-characterized folding pathway of the Tetrahymena LSU group I intron, it was shown that Cyt-19 acts by disrupting a nonnative secondary structure that acts as a kinetic trap in the folding of this intron. Binding of Cyt-18 to the intron RNA already greatly improves the stability of the RNA, but the reaction does not go to completion, with a significant percentage of the population remaining trapped in misfolded conformations. Recruitment of the Cyt-19 RNA helicase leads to resolution of these kinetic traps, and folding proceeds to completion.
Hfq, A Protein That Helps RNA Molecules to Anneal
RNA antisense approaches in research or in therapy are often hampered by the low accessibility of target RNA regions due to local secondary structure, even in supposedly unstructured mRNAs. This problem is also encountered by some E. coli noncoding RNAs (ncRNAs), which are a class of naturally occurring antisense RNAs that function in post-transcriptional regulation. Recently, genome-wide screens have brought the number of small RNAs in E. coli to around 50, and a large number of these RNAs may regulate post-transcriptional gene expression. 11 These RNAs fulfill their function by partial base pairing to target mRNA transcripts, thereby blocking ribosome access to the Shine-Dalgarno sequence; by opening up inhibitory mRNA structures; or by influencing mRNA stability. The conformational switches in the 5'-UTR of mRNAs in these cases are induced by ncRNAs.
NcRNAs and their targets often show discrete structures in regions that later must anneal with the interaction partner. Whereas the structure of ncRNAs and their target RNAs may have evolved to facilitate their mutual interaction, it is believed that protein factors enhance these RNA-RNA interactions. A candidate for this activity in E. coli is the protein Hfq, which was first described as a cofactor required for replication of the RNA phage Qβ.33 Hfq is strikingly conserved in a wide range of bacteria and highly abundant.34 It has been estimated that there are approximately 30,000 to 60,000 Hfq molecules per bacterial cell.35 Hfq has been shown to interact with, or to be required for the activity of, several small interfering RNAs, including DsrA,36 RprA,37 OxyS,38,39 Spot4234 and RyhB.40
The Hfq-dependent action of the small RNAs RyhB and OxyS is particularly enlightening with regard to the mechanism of Hfq activity on these RNAs (fig. 3). RyhB regulates superoxide dismutase B (sodB) mRNA translation and stability. SODs eliminate free superoxide radicals and are therefore key enzymes of the cellular defense system against oxidative stress. The RyhB-sodB mRNA interaction requires the presence of Hfq and occurs over a stretch of nine complementary nucleotides, encompassing the AUG initiation codon of translation on the sodB mRNA. RyhB binding blocks the translation initiation codon of sodB and triggers the RNase E dependent degradation of both RyhB and sodB mRNA.41 In this system, it is the mRNA and not the small regulatory RNA that is bound by Hfq with high affinity.42 Hfq binds to sodB mRNA with a dissociation constant of Kd = 1.8 nM. Hfq binds to RyhB much less strongly with a Kd = 1.5 μM, which is the same order of magnitude as that for binding of Hfq to DsrA, OxyS or Spot42 RNA.34,36,39,43
Because the affinity of Hfq for RyhB RNA is almost three orders of magnitude lower than for sodB mRNA, it is interesting to look at these interactions at the structural level. The Hfq binding site on sodB mRNA is an A/U rich single stranded linker of 14 nucleotides between two stem-loops; the first stem-loop (stem-loop a) begins with the first transcribed nucleotide (fig. 3). The second stem-loop (stem-loop b) encompasses the region in which translation is initiated. Following addition of Hfq, the A/U-rich region is protected against RNAse E cleavage, indicating that Hfq binds to and protects this region. In strong contrast, addition of Hfq changes the structure of the stem-loop b region, rendering it more accessible to RyhB. Since this region is complementary to the RyhB RNA, Hfq facilitates binding of RyhB to sodB mRNA by resolving a base-pairing interaction that interferes with binding of RyhB.
In a similar way, Hfq mediates interaction of fhlA mRNA and the small RNA OxyS. OxyS RNA is a 109-nucleotide untranslated RNA that is induced in response to oxidative stress in E. coli and activates or represses multiple genes.44 Among the repressed genes are rpoS and the transcriptional activator fhlA. OxyS repression of fhlA is achieved through two base-pairing interactions, one site that overlaps with the ribosome-binding site and the second site that resides in the fhlA mRNA coding sequence.45,46 OxyS binding prevents ribosome access to the fhlA mRNA and leads to the repression of translation. Hfq was shown to increase OxyS interaction with its target mRNAs by formation of supershifted complexes that did not depend on the continued presence of Hfq, which was digested away by proteinase K.39 OxyS RNA consists of three stem-loops and a single-stranded A/U rich region. While some positions became more protected upon Hfq binding, mainly in the single stranded region downstream of stem-loop b, some positions outside the binding site showed increased accessibility. Thus, Hfq seems to function by increasing the accessibility of an RNA sequence that is required for interaction with a target RNA.
Interestingly, Hfq shows some characteristic features of eukaryotic Sm proteins, which consist of a group of seven proteins that form a heteroheptameric ring structure and bind to U-rich sequences of the spliceosomal U RNAs. Hfq has in common with the Sm proteins t presence of the Sm1 motif, formation of an oligomeric ring structure, and preferential binding to poly-U RNA. Crystal structures of the Staphylococcus aureus Hfq and of a complex of Hfq bound to the oligoribonucleotide AUUUUUG provide some insight into the molecular details of Hfq action.47 Hfq forms a symmetric hexameric ring with a diameter of ˜65 Å and a width of ˜23 Å (fig. 3B). The hexamer has a doughnut shape with a central hole of ˜12 Å diameter. The Hfq-RNA structure reveals that the single-stranded 7-mer oligoribonucleotide binds in a circularly unwound conformation around the central basic cleft in the pore of one face of the Hfq hexamer. The structural data explain the preferential binding of Hfq to A/U rich RNA and also provide insight into the mode of action of Hfq on RNA-RNA interactions: when Hfq binds single-stranded RNA, the RNA is unwound within its central pore. The structure indicates that Hfq could accommodate A/U-rich RNA for up to six nucleotides. Such binding and unwinding strongly destabilizes surrounding RNA structures that are located several nucleotides on either side of the binding site, thereby permitting new RNA-RNA interactions.
Proteins with RNA Chaperone Activity
RNA molecules may fold into nonnative but stable structures that lie along the folding pathways to the native conformation.32,48-51 These alternative structures represent kinetic traps; they have long lifetimes due to the energy required to break the incorrect interactions. RNA duplexes have a very high thermodynamic stability. An RNA helix of 10 base pairs (bp) in length has a dissociation half-life of about 30 min; G/C-rich 10-bp duplexes might have dissociation halftimes of up to 100 years at 30°C.52,53 Alternative folding states already exist in relatively small RNA molecules like tRNAs. For tRNALeu it has been shown that an inactive conformation is stable on the hours timescale and that it can be converted to an active structure upon heating in the presence of Mg2+.54
How can RNA overcome this folding problem and reach its active, native conformation? One possible solution is the existence of proteins that help folding. Nonspecific RNA-binding proteins solve the kinetic folding problem by acting as RNA chaperones. RNA chaperones are defined as proteins that prevent RNA misfolding and/or resolve misfolded RNAs, thus enabling correct folding of the RNA molecule.53 The idea of proteins that aid RNA in the folding process was indicated over 20 years ago, when it was shown that a fragment of hnRNP A1 protein can renature kinetically trapped 5S and tRNAs.55,56 RNA chaperones are a highly diverse family of nucleic acid-binding proteins that exhibit a wide variety of biological activities. No common structural signature has been identified, because they belong to different classes of RNA-binding proteins. In many cases, no known RNA binding domain can be predicted.57 RNA chaperones seem to recognize neither a common sequence nor a common structural motif.58-62
An increasing number of proteins have been shown to act as RNA chaperones. In vitro, RNA chaperone activity can be monitored in different ways. Assays include the measurement of RNA annealing activity, strand exchange activity, and ribozyme turnover stimulation, as well as the promotion of trans-splicing of the T4 td intron.13,14,63-65 Taking advantage of a misfolded species of the T4 phage td pre-mRNA in the absence of translation, an in vivo assay for RNA chaperone activity was established. Using this assay, it was possible to show that a series of proteins with in vitro RNA chaperone activity can also resolve a misfolded RNA structure in vivo. The most prominent of these RNA chaperones is the E. coli protein StpA.66-68
E. coli Protein StpA
StpA was initially isolated as a multicopy suppressor of a splicing defective mutant of the bacteriophage T4 td group I intron.69 It is a 15.3-kDa nucleoid-associated protein present in Escherichia, Shigella and Salmonella species and shares 58% sequence identity with its E. coli paralogue H-NS.69,70 StpA has a nucleic acid binding domain in its C-terminal region, whereas its N-terminus functions as dimerization domain.71 It was shown that StpA enhances splicing in vitro by using a trans-splicing system, where the td intron is split into two half-molecules. RNA strand-annealing reactions can be greatly enhanced by adding StpA. By means of strand exchange assays, it could be shown that StpA also enhances RNA dissociation, a property required to resolve misfolded structures. StpA is dispensable after assembly of the trans-splicing precursors, because removing the protein with proteinase K treatment before initiating catalysis does not affect the level of splicing enhancement. This demonstrates that StpA is not needed for catalysis, but only for the assembly of the catalytically active group I intron conformation.14 StpA also induces strand transfer during primer extension by the HIV reverse transcriptase.72
In vivo, StpA rescues folding of the td pre-mRNA in mutants that are kinetically trapped in nonactive conformations due to the absence of translation.66,67 In vivo DMS modification of the T4 td group I intron RNA showed that in presence of StpA, tertiary structure elements become more accessible to the modifying agent, suggesting that the protein has a destabilizing effect on the overall tertiary structure of the intron. This suggests that the RNA chaperone activity of StpA results partly from the resolution of tertiary interactions in the intron structure. This activity may be a general characteristic of proteins with RNA chaperone activity, but this still remains to be analyzed.73
Nucleocapsid Protein of HIV-1
Retroviral nucleocapsid (NC) proteins are examples of viral proteins that exert RNA chaperone activities for several RNA switches during viral replication and assembly. HIV-1 NC is a highly basic protein consisting of 55 amino acids and two zinc finger structures. The nucleic acid chaperone activity of NC greatly accelerates tRNALys annealing to the primer-binding site, and it facilitates minus- and plus-strand transfer reactions of retroviral reverse transcriptase.74,75 The zinc fingers are not essential for tRNALys annealing, but they are responsible for inducing subtle tertiary structure changes and for destabilizing helical regions of the tRNA.76 In contrast, the zinc finger structures are critical for the annealing step in minus-strand transfer.77
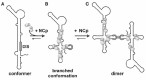
The HIV-1 genome is packaged as a dimer into a viral particle. The two copies of the RNA are linked through base pairing of the dimer linkage sites (DLS) located in the 5'-leader of the RNA. Studies on the overall structure of the full length HIV-1 leader RNA have revealed the existence of a compactly folded structure of the RNA induced by base-pairing interactions between sequences that would otherwise form the poly-A and the dimerization initiation site (DIS) domains.78-83 This compact structure masks the DIS loop, and the RNA is in a dimerization-incompetent form. Before the dimerization of the genome, the compact structure is lost and a branched structure containing the poly-A and DIS domains is formed, suggesting that this conformational switch regulates HIV dimerization (fig. 4). The viral NC protein has been shown to stimulate dimerization of HIV-1 RNA at physiological conditions.84,85 Using band-shift assays, it could be shown that upon addition of NC protein, the compact structure is disrupted and dimerization is enabled through exposure of the DIS hairpin, which indicates that NC enhances dimerization through stabilization of the branched structure. It favors the branched folding over the thermodynamically more stable rod-like conformer structure of the HIV-1 leader RNA. The dimerization reaction is initiated by base pairing of the self-complementary DIS loops, forming a kissing complex. NC accelerates the transition of the kissing complex into the more stable extended complex.86-88 The existence of two different structures of the HIV-1 RNA creates the possibility for the leader RNA to function as a regulatory switch during viral replication. The structural transition initiated by the NC protein may lead to the formation of signals necessary for RNA dimerization, packaging, and also reverse transcription.89
The general and nonspecific nature of NC's RNA chaperone activity was further demonstrated through its acceleration of ribozyme activity. NC can facilitate both the formation and disruption of interactions between the hammerhead ribozyme and its substrates and products, thus increasing the rate of catalysis. The protein also resolves kinetically trapped misfolded complexes of the ribozyme.13,62,64,90
Proteins that Help with the Formation of RNA-Protein Complexes
One of the most prominent RNA switches known to date is the formation of the U2/U6 snRNP complex from the U4/U6. The spliceosome is highly dynamic, with multiple RNA-RNA and RNA-protein rearrangements occurring during assembly and disassembly. Little is known about how these switches are induced, but we can look at the proteins that build up the snRNPs and those required for its assembly. The spliceosomal snRNPs U1, U2, U4 and U5 contain a common RNP structure termed the Sm core formed by the binding of Sm proteins onto the U snRNAs. The Sm proteins are reminiscent of Hfq, which is “Sm-like”, in that they form a ring-shaped multimeric structure. In vitro, the Sm proteins bind spontaneously to the snRNAs, but in vivo an additional complex was found to be necessary for the assembly of the spliceosomal core complexes. The SMN complex is named after the “survival of motor neurons” protein implicated in spinal muscular atrophy disease. Reduced expression of SMN results in degeneration of motor neurons and weakness of voluntary muscles.91 SMN is part of a complex involved in the assembly of the Sm core,92 which probably facilitates binding of the Sm proteins to the snRNAs. The Sm proteins are modified through symmetrical arginine dimethylation, enhancing the binding of Sm proteins to the SMN complex.
In yeast, no equivalent for the SMN complex has been found. Instead, the La auto antigen might be involved in the facilitation of snRNP assembly. Whether La can functionally replace the SMN protein remains to be tested. The first protein that binds to newly transcribed RNA polymerase III transcripts is the highly conserved La auto antigen. In yeast, this protein, called Lhp1p, is required for tRNA maturation.93 Another role for La might also be assisting in the assembly of the U6 snRNP. U6 is an RNA polymerase III transcript and in contrast to the other spliceosomal RNAs, U6 is not bound by the Sm proteins, but instead by a set of seven Sm-like proteins, the Lsm proteins.94 A mutation in the Lsm8p protein results in a reduced level of mature U6 snRNPs, consistent with a defect in U6 snRNP assembly. The exciting fact about this mutant is that it requires the La protein Lhl1p for growth, suggesting that La acts as a molecular chaperone for U6 snRNP assembly.95 The La protein might resemble Hfq in its mode of action in that it binds a set of RNAs that are relatively structured, thereby facilitating their assembly with other RNAs or proteins. The way in which La chaperones the folding process of its RNA targets is unknown.
Perspective
Our understanding of how proteins modulate RNA structures has improved substantially in the past few years. Experiments have revealed a large variety of interaction modes, both in specificity and in the way that proteins influence RNA folding. The number of well-characterized RNA-protein complexes is increasing steadily, but detailed mechanistic studies have been reported for some examples. We expect that kinetic and structural analyses of these interactions will reveal as-yet unknown activities of proteins on RNA structure, folding, and function. Studies of dynamic interactions will shed light on the mechanisms involved in the induction of RNA structural changes.
Acknowledgements
We thank all the members of the Schroeder lab for comments and critical reviewing of the manuscript and especially Dr. Paul Watson for the help with English grammar and references. Work in our laboratory is funded by the Austrian Science Fund (FWF) grants F1703, F1704, P16026 and Z72.
References
- 1.
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. [PubMed: 9241415]
- 2.
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244(4900):48–52. [PubMed: 2468181]
- 3.
- Schultes EA, Bartel DP. One sequence, two ribozymes: Implications for the emergence of new ribozyme folds. Science. 2000;289(5478):448–452. [PubMed: 10903205]
- 4.
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289(5800):751–758. [PubMed: 7007895]
- 5.
- Nahvi A, Sudarsan N, Ebert MS. et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. [PubMed: 12323379]
- 6.
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419(6910):952–956. [PubMed: 12410317]
- 7.
- Winkler WC, Nahvi A, Roth A. et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428(6980):281–286. [PubMed: 15029187]
- 8.
- Mandal M, Boese B, Barrick JE. et al. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113(5):577–586. [PubMed: 12787499]
- 9.
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74(3):475–482. [PubMed: 8348614]
- 10.
- Henkin TM. Control of transcription termination in prokaryotes. Annu Rev Genet. 1996;30:35–57. [PubMed: 8982448]
- 11.
- Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31(7):1813–1820. [PMC free article: PMC152812] [PubMed: 12654996]
- 12.
- Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109(6):769–779. [PubMed: 12086675]
- 13.
- Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262(5130):99–102. [PubMed: 7692597]
- 14.
- Zhang A, Derbyshire V, Salvo JL. et al. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1(8):783–793. [PMC free article: PMC1369319] [PubMed: 7493324]
- 15.
- Caprara MG, Lehnert V, Lambowitz AM. et al. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell. 1996;87(6):1135–1145. [PubMed: 8978617]
- 16.
- Solem A, Chatterjee P, Caprara MG. A novel mechanism for protein-assisted group I intron splicing. RNA. 2002;8(4):412–425. [PMC free article: PMC1370265] [PubMed: 11991637]
- 17.
- Buchmueller KL, Webb AE, Richardson DA. et al. A collapsed nonnative RNA folding state. Nat Struct Biol. 2000;7(5):362–366. [PubMed: 10802730]
- 18.
- Buchmueller KL, Weeks KM. Near native structure in an RNA collapsed state. Biochemistry. 2003;42(47):13869–13878. [PubMed: 14636054]
- 19.
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24(5):192–198. [PubMed: 10322435]
- 20.
- Tanner NK, Linder P. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol Cell. 2001;8(2):251–262. [PubMed: 11545728]
- 21.
- Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat Struct Biol. 2001;8(2):113–116. [PubMed: 11175897]
- 22.
- Silverman E, Edwalds-Gilbert G, Lin R. DExD/H-box proteins and their partners: Helping RNA hilecases unwind. Gene. 2003;312:1–16. [PubMed: 12909336]
- 23.
- Linder P, Lasko PF, Ashburner M. et al. Birth of the D-E-A-D box. Nature. 1989;337(6203):121–122. [PubMed: 2563148]
- 24.
- Gorbalenya AE, Koonin EV. Helicases: Amino-acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429.
- 25.
- Jankowsky E, Gross CH, Shuman S. et al. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291(5501):121–125. [PubMed: 11141562]
- 26.
- Rocak S, Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5(3):232–241. [PubMed: 14991003]
- 27.
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. [PubMed: 10872469]
- 28.
- Rogers JrGW, Komar AA, Merrick WC. eIF4A: The godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. [PubMed: 12206455]
- 29.
- Linder P, Stutz F. mRNA export: Travelling with DEAD box proteins. Curr Biol. 2001;11(23):R961–963. [PubMed: 11728322]
- 30.
- Korolev S, Yao N, Lohman TM. et al. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7(3):605–610. [PMC free article: PMC2143965] [PubMed: 9541392]
- 31.
- Grainger RJ, Norman DG, Lilley DM. Binding of U1A protein to the 3' untranslated region of its premRNA. J Mol Biol. 1999;288(4):585–594. [PubMed: 10329165]
- 32.
- Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109(7):797–800. [PubMed: 12110176]
- 33.
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219(154):588–590. [PubMed: 4874917]
- 34.
- Moller T, Franch T, Hojrup P. et al. Hfq. A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9(1):23–30. [PubMed: 11804583]
- 35.
- Kajitani M, Kato A, Wada A. et al. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol. 1994;176(2):531–534. [PMC free article: PMC205081] [PubMed: 8288550]
- 36.
- Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183(6):1997–2005. [PMC free article: PMC95095] [PubMed: 11222598]
- 37.
- Wassarman KM, Repoila F, Rosenow C. et al. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15(13):1637–1651. [PMC free article: PMC312727] [PubMed: 11445539]
- 38.
- Zhang A, Altuvia S, Tiwari A. et al. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17(20):6061–6068. [PMC free article: PMC1170932] [PubMed: 9774349]
- 39.
- Zhang A, Wassarman KM, Ortega J. et al. The Sm-like Hfq protein increases Oxys RNA interaction with target mRNAs. Mol Cell. 2002;9(1):11–22. [PubMed: 11804582]
- 40.
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99(7):4620–4625. [PMC free article: PMC123697] [PubMed: 11917098]
- 41.
- Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17(19):2374–2383. [PMC free article: PMC218075] [PubMed: 12975324]
- 42.
- Geissmann TA, Touati D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. [PMC free article: PMC1271764] [PubMed: 14739933]
- 43.
- Brescia CC, Mikulecky PJ, Feig AL. et al. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9(1):33–43. [PMC free article: PMC1370368] [PubMed: 12554874]
- 44.
- Altuvia S, Weinstein-Fischer D, Zhang A. et al. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell. 1997;90(1):43–53. [PubMed: 9230301]
- 45.
- Altuvia S, Zhang A, Argaman L. et al. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17(20):6069–6075. [PMC free article: PMC1170933] [PubMed: 9774350]
- 46.
- Argaman L, Altuvia S. fhlA repression by OxyS RNA: Kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol. 2000;300(5):1101–1112. [PubMed: 10903857]
- 47.
- Schumacher MA, Pearson RF, Moller T. et al. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21(13):3546–3556. [PMC free article: PMC126077] [PubMed: 12093755]
- 48.
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270(36):20871–20874. [PubMed: 7545662]
- 49.
- Treiber DK, Williamson JR. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J Mol Biol. 2001;305(1):11–21. [PubMed: 11114243]
- 50.
- Woodson SA. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci. 2000;57(5):796–808. [PubMed: 10892344]
- 51.
- Thirumalai D, Lee N, Woodson SA. et al. Early events in RNA folding. Annu Rev Phys Chem. 2001;52:751–762. [PubMed: 11326079]
- 52.
- Turner DH, Sugimoto N, Freier SM. Thermodynamics and kinetics of base-pairing and of DNA and RNA self-assembly and helix coil transition: Springer-Verlag. 1990 .
- 53.
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270(36):20871–20874. [PubMed: 7545662]
- 54.
- Lindahl T, Adams A. Native and renatured transfer ribonucleic acid. Science. 1966;152(721):512–514. [PubMed: 5910188]
- 55.
- Karpel RL, Burchard AC. Physical studies of the interaction of a calf thymus helix-destablizing protein with nucleic acids. Biochemistry. 1980;19(20):4674–4682. [PubMed: 6252954]
- 56.
- Karpel RL, Miller NS, Fresco JR. Mechanistic studies of ribonucleic acid renaturation by a helix-destabilizing protein. Biochemistry. 1982;21(9):2102–2108. [PubMed: 6178431]
- 57.
- Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002;72:223–268. [PubMed: 12206453]
- 58.
- Munroe SH, Dong XF. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA-RNA annealing. Proc Natl Acad Sci USA. 1992;89(3):895–899. [PMC free article: PMC48351] [PubMed: 1371011]
- 59.
- Portman DS, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13(1):213–221. [PMC free article: PMC394795] [PubMed: 7508381]
- 60.
- Darlix JL, Lapadat-Tapolsky M, de Rocquigny H. et al. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254(4):523–537. [PubMed: 7500330]
- 61.
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7(3):336–342. [PubMed: 9204274]
- 62.
- Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem Sci. 1998;23(8):297–301. [PubMed: 9757830]
- 63.
- Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8(13):1575–1588. [PubMed: 7958841]
- 64.
- Herschlag D, Khosla M, Tsuchihashi Z. et al. An RNA chaperone activity of nonspecific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13(12):2913–2924. [PMC free article: PMC395173] [PubMed: 8026476]
- 65.
- Nedbal W, Frey M, Willemann B. et al. Mechanistic insights into p53-promoted RNA-RNA annealing. J Mol Biol. 1997;266(4):677–687. [PubMed: 9102461]
- 66.
- Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12(9):1327–1337. [PMC free article: PMC316773] [PubMed: 9573049]
- 67.
- Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18(13):3776–3782. [PMC free article: PMC1171454] [PubMed: 10393192]
- 68.
- Moll I, Leitsch D, Steinhauser T. et al. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4(3):284–289. [PMC free article: PMC1315900] [PubMed: 12634847]
- 69.
- Zhang A, Belfort M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20(24):6735. [PMC free article: PMC334595] [PubMed: 1480493]
- 70.
- Zhang A, Rimsky S, Reaban ME. et al. Escherichia coli protein analogs StpA and H-NS: Regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15(6):1340–1349. [PMC free article: PMC450038] [PubMed: 8635467]
- 71.
- Cusick ME, Belfort M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol Microbiol. 1998;28(4):847–857. [PubMed: 9643551]
- 72.
- Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: Reshuffling of genetic markers mediated by RNA chaperones. Proc Natl Acad Sci USA. 2000;97(12):6385–6390. [PMC free article: PMC18612] [PubMed: 10829081]
- 73.
- Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. [PMC free article: PMC186668] [PubMed: 12208852]
- 74.
- De RocquignyH, Gabus C, Vincent A. et al. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89(14):6472–6476. [PMC free article: PMC49523] [PubMed: 1631144]
- 75.
- Allain B, Lapadat-Tapolsky M, Berlioz C. et al. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13(4):973–981. [PMC free article: PMC394899] [PubMed: 7509280]
- 76.
- Hargittai MR, Mangla AT, Gorelick RJ. et al. HIV-1 nucleocapsid protein zinc finger structures induce tRNA(Lys,3) structural changes but are not critical for primer/template annealing. J Mol Biol. 2001;312(5):985–997. [PubMed: 11580244]
- 77.
- Guo J, Wu T, Anderson J. et al. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol. 2000;74(19):8980–8988. [PMC free article: PMC102094] [PubMed: 10982342]
- 78.
- Berkhout B, van WamelJL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6(2):282–295. [PMC free article: PMC1369913] [PubMed: 10688366]
- 79.
- Berkhout B, Klaver B, Das AT. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207(1):276–281. [PubMed: 7755727]
- 80.
- Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5' major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33(45):13464–13474. [PubMed: 7947755]
- 81.
- Skripkin E, Paillart JC, Marquet R. et al. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. [PMC free article: PMC43906] [PubMed: 8197162]
- 82.
- Mujeeb A, Clever JL, Billeci TM. et al. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5(6):432–436. [PubMed: 9628479]
- 83.
- Girard F, Barbault F, Gouyette C. et al. Dimer initiation sequence of HIV-1Lai genomic RNA: NMR solution structure of the extended duplex. J Biomol Struct Dyn. 1999;16(6):1145–1157. [PubMed: 10447199]
- 84.
- Darlix JL, Gabus C, Nugeyre MT. et al. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216(3):689–699. [PubMed: 2124274]
- 85.
- Muriaux D, Girard PM, Bonnet-Mathoniere B. et al. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5' leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270(14):8209–8216. [PubMed: 7713927]
- 86.
- Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35(5):1589–1598. [PubMed: 8634290]
- 87.
- Paillart JC, Marquet R, Skripkin E. et al. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie. 1996;78(7):639–653. [PubMed: 8955907]
- 88.
- Windbichler N, Werner M, Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: Coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31(22):6419–6427. [PMC free article: PMC275567] [PubMed: 14602899]
- 89.
- Huthoff H, Berkhout B. Two Alternative strcutures of the HIV-1 leader RNA. RNA. 2001;7:143–157. [PMC free article: PMC1370064] [PubMed: 11214176]
- 90.
- Bertrand EL, Rossi JJ. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13(12):2904–2912. [PMC free article: PMC395172] [PubMed: 8026475]
- 91.
- Crawford TO. From enigmatic to problematic: The new molecular genetics of childhood spinal muscular atrophy. Neurology. 1996;46(2):335–340. [PubMed: 8614490]
- 92.
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90(6):1023–1029. [PubMed: 9323130]
- 93.
- Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375–403. [PubMed: 12045101]
- 94.
- Mayes AE, Verdone L, Legrain P. et al. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18(15):4321–4331. [PMC free article: PMC1171508] [PubMed: 10428970]
- 95.
- Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;15:7442–74453. [PMC free article: PMC1171088] [PubMed: 9857199]
- Introduction
- Proteins That Bind RNA with High Affinity and Stabilize Specific Structures
- RNA Helicases
- NPH-II
- Cyt-19
- Hfq, A Protein That Helps RNA Molecules to Anneal
- Proteins with RNA Chaperone Activity
- Nucleocapsid Protein of HIV-1
- Proteins that Help with the Formation of RNA-Protein Complexes
- Perspective
- Acknowledgements
- References
- Protein-Induced RNA Switches in Nature - Madame Curie Bioscience DatabaseProtein-Induced RNA Switches in Nature - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...