NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Although many known receptors that regulate addiction have been pharmacologically and biochemically well characterized, some orphan receptors with homology to known receptors of abuse (i.e. GPR35) remain uncharacterized. GPR35 is a G-protein coupled receptor, first identified in 1998 after a screen of a human genomic library. More recent RT-PCR studies have now confirmed the presence of GPR35 in dorsal root ganglion, the cerebellum and brain, as well as GPR35b, which was cloned from a human whole brain cDNA library. Thus, GPR35 regulation appears to have profound physiological and pathophysiological implications. In line with the specific aim of this project, the identified probes ML145 (CID-2286812) and ML144 (CID-1542103) represent selective antagonists for GPR35, but not for the related GPR55 orphan receptor, supporting the hypothesis that they do not produce non-specific interference with signaling directly at or downstream of the β-arrestin signaling pathway. These probes will serve as novel tools to delineate the biochemistry of GPR35 as potential therapeutics to selectively target pathways underlying pain and to enhance our understanding of the molecular basis of addiction.
Probe project: Selective GPR35 Antagonists
Assigned Assay Grant #: 1 X01 MH085708-01
Screening Center Name & PI: Sanford-Burnham Center for Chemical Genomics (NIH PubChem & MLPCN designation) & John C. Reed
Chemistry Center Name & PI: Burnham Center for Chemical Genomics & John C. Reed
Assay Submitter & Institution: Lawrence S. Barak, Duke University Medical Center Collaborating PI: (Mary E. Abood, Temple University, PA)
PubChem Summary Bioassay Identifier (AID): AID-2079
Probe(s) Structure & Characteristics
This Center Probe Report describes two selective antagonists for GPR35 an orphan GPCR receptor that represent two novel scaffolds or chemical series: (1) CID2286812 and(2) CID1542103


| CID | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Selectivity | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID2286812 (scaffold1) ML145 | GPR35 Antagonist Orphan GPCR receptor | 20.1 nM SID-87544499 AID-2480 | GPR55 Antagonist Orphan GPCR receptor | ~ 21.7 µM SID-87544499 AID-2397 | ~1,080-fold vs. GPR55 Antagonist | N/A |
| CID1542103 (scaffold2) ML144 | 2220 nM SID-87544496 AID-2480 | >32 µM SID-87544496 AID-2397 | >15-fold vs. GPR55 Antagonist | N/A |
Recommendations for the scientific use of this probe
Many receptors regulating addiction are pharmacologically and biochemically well characterized, but some orphan receptors like GPR35 with homology to known receptors of abuse remain uncharacterized. The aim of this research is to identify small molecule antagonists of human GPR35. The identification of small molecules capable of selectively inhibiting or activating orphans will provide tools for elucidating novel molecular pathways underlying addictive behaviors. These novel compounds will then be utilized to elucidate a number of things such as characterize GPR35 biology in vitro, GPR35 in animal models of pain and enhance the understanding of the molecular basis of addiction.
1. Scientific Rationale for Project
Drug addiction continues to remain a major public health concern in the United States. Addictive behavior results from changes in central nervous system signaling pathways that are modified after exposure to drugs of abuse. In particular, compounds such as cannabinoids and opiates that influence mood and pain perception are commonly associated with addictive behaviors. Many receptors regulating addiction are pharmacologically and biochemically well characterized, but some orphan receptors like GPR35 with homology to known receptors of abuse remain almost totally uncharacterized. GPR35 is a G-protein coupled receptor that was first identified in 1998 after a screen of a human genomic library (1). GPR35 is homologous to P2Y purinergic receptors and GPR23, whose ligand is lysophosphatidic acid. GPR35 shares a 30 percent identity with the putative cannabinoid receptor GPR55 (2–5). The ability of GPR55 to recognize cannabinoids was first described in a yeast expression system, where the CB1 antagonists AM251 and SR141716A (rimonabant) acted as agonists (6). Preliminary studies of GPR35 by mRNA expression showed it expressed predominantly in the immune and gastrointestinal systems (1). However, recent RT-PCR studies have confirmed the presence of GPR35 in dorsal root ganglion, the cerebellum and brain, and GPR35b was cloned from a human whole brain cDNA library (2, 5, 7). Variable Gi/o protein activation by GPR35 that was pertussis toxin sensitive was subsequently observed in rat sympathetic neurons (2).
There are approximately fifteen papers characterizing GPR35 in the PubMed listed peer reviewed literature. An N-terminal splice variant of GPR35, GPR35b, was identified from a genetic screen of gastric carcinomas (8), leading to speculation that GPR-35 regulates cell growth. The observation that the a isoform possessed a stronger transforming activity than the b also led the authors to postulate that GPR-35a possesses constitutive activity (8). While GPR35 has been implicated in the formation of gastric cancers (8), conversely, deletion of GPR35 may be responsible for a mental retardation syndrome associated with deletions on 2q37.3 (9).
GPR35 regulation appears to have profound physiological and pathophysiological implications so that defining compounds that regulate GPR35 will be important. The specific aim of this grant is to identify small molecule antagonists of human GPR35. These novel compounds will be utilized to characterize GPR35 biology in vitro and GPR35 in animal models of pain. Thus, this proposal will provide tools for delineating the biochemistry of GPR35, potentially provide compounds for targeted therapeutics of pathways underlying pain, and enhance our understanding of the molecular basis of addiction.
2. Project Description
a. Original goal for probe characteristics
The goal of the HTS was to identify novel and specific inhibitors of GPR35. To date, no antagonists for GPR35 are known and the goal of this project was to identify small molecules that had an IC50 of 5 µM or less in the primary GPR35 β-arrestin HCS assay, with at least 10-fold antagonist selectivity against the related receptor GPR55.
b. Information for each Assay Implemented and Screening Run
i. PubChem Bioassay Name(s), AID(s), Assay-Type (Primary, DR, Counterscreen, Secondary)
Table 1Assays for GPR35
| PubChem BioAssay Name | AID | Probe Type | Assay Type | Assay Format | Assay Detection (well format) |
|---|---|---|---|---|---|
| Summary of Image-based HTS for Selective Antagonists of GPR35 | 2079 | N/A | Summary | N/A | N/A |
| Image-Based HTS for Selective Antagonists of GPR35 [Primary] | 2058 | Inhibitors | Primary | Cell-based | Imaging method (384) |
| HCS GPR35 Antagonist SAR-primary assay used as secondary | 2480 | Inhibitors | SAR | Cell-based | Imaging method (384) |
| HCS GPR55 antagonist – Counterscreen - SAR | 2397 | Inhibitors | SAR | Cell-based | Imaging method (384) |
ii. Assay Rationale & Description
Primary Screen
This image-based high-content screen (HCS) is based on fluorescence redistribution of a GFP-β-arrestin complex from homogeneous distribution in the cytoplasm via the plasma membrane to intracellular pits and vesicles (assay technology marketed as Transfluor® assay by Molecular Devices). Upon activation by ligand binding, GPCRs undergo deactivation or “desensitization” by binding of the β-arrestin protein to the activated receptor. The GPCR-β-arrestin complex internalizes, the ligand is removed and the receptor is recycled back to the cell membrane. Localization of the fluorescently labeled β-arrestin can be monitored by image analysis (10). The primary screen assay is designed to identify compounds inhibiting GPR35 signaling induced by the GPR35 agonist Zaprinast at approx. EC80 concentration (5).
The primary screening protocol is described below.
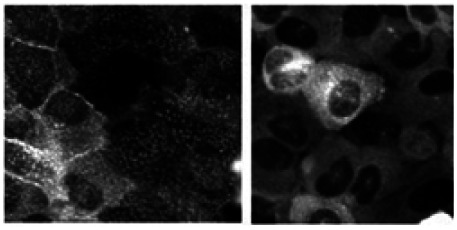
GPR35 Example Images of Positive and Negative Controls
Effect of 10 µM agonist Zaprinast (left panel) compared to DMSO control (right panel)
Primary Assay Materials
Table 2Critical Reagents used for the uHTS experiments
| Reagent | Vendor |
|---|---|
| U2OS (Human Osteosarcoma) cell line stably expressing GFP-tagged β-arrestin and over-expressing the GPR35 receptor | Cells from AP, scaled-up by BCCG |
| Zaprinast - GPR35 Agonist | ALEXIS Biochemicals (now Enzo Life Sciences) |
| Paraformaldehyde - Fixative | ACROS Organics |
| DAPI – Nuclei Stain | Invitrogen |
Primary Screen Protocol
A. Plate Preparation:
- 45ul of cell suspension (133,000 cells/ml in culture medium) was dispensed in each well of the assay plates using a Wellmate bulk dispenser.
- Plates were incubated overnight or approximately 20 hours at 37 degree C and 5% CO2.
- Serum was removed by media aspiration and replaced with 45ul serum-free MEM prior to addition of compounds.
- Compound addition was done on a Biomek FX with 384-head dispenser (Beckman):
- 5ul of 100µM compound solution was added to columns 3 through 24 of the assay plates for a final assay compound concentration of 10µM and 0.5% DMSO.
- 5ul of 5% DMSO was added to columns 1 and 2 to balance the volume and DMSO concentration across the plate.
- 5ul of positive control (2% DMSO) working solution was added to column 1.
- Plates were incubated for 15 minutes at room temperature.
- Agonist addition was done on a Biomek FX with 384-head dispenser (Beckman). 5ul of agonist (100µM Zaprinast) working solution was added to columns 2–24. (This also serves as the negative control in column 2.)
- Plates were incubated for 45 minutes at 37 degrees C and 5% CO2.
- Media was aspirated leaving 20ul liquid in each well using a Titertek plate washer.
- 40ul of fixative working solution was added to each well using a Wellmate bulk dispenser (Matrix) for a final concentration of 4% PFA.
- Plates were incubated for 40 minutes at room temperature.
- Fixative was aspirated and plates were washed twice with 50ul PBS leaving 20ul liquid in each well using a Titertek plate washer.
- 40ul of DAPI working solution was added using a Wellmate bulk dispenser for a final DAPI concentration of 100ng/ml. Aluminum plate seals were applied to each plate.
B. Image Acquisition and Analysis:
- Image acquisition was performed on an Opera QEHS (Perkin Elmer) with 45 plate capacity loader/stacker and the following settings:
- -
40x 0.6 NA air objective
- -
Acquisition camera set to 2-by-2 binning for an image size of 688 by 512 pixels
- -
2 channels acquired sequentially: Exp1Cam1 = B-arrestin GFP using 488 nm laser excitation
and 540/70 nm 4mission filters, Exp2Cam2 = DAPI (nuclei) using 365 nm Xenon lamp excitation and 450/50 nm emission filters
- -
3 fields per well
- Image analysis was performed using the Acapella™ Spot Detection Algorithm with the following analysis settings:
NUCLEI DETECTION - Threshold Adjustment: 4 - Nuclear Splitting Adjustment: 10 - Individual Threshold Adjustment 0.05 - Minimum Nuclear Area: 200 - Minimum Nuclear Contrast: 0 CYTOPLASM DETECTION - Cytoplasm Individual Threshold Adjustment: 0 SPOT DETECTION - Spot Minimum Distance 3 - Spot Peak Radius 0 - Spot Reference Radius 3 - Spot Minimum Contrast 0.26 - Spot Minimum to Cell Intensity 0.5 - Metrics calculated from…NUCLEI IMAGES: Cell Count (“Number of Cells Analyzed”), Nuclei Area (“Area of the Nucleus”), Integrated Intensity of the Nuclei (“Total Integrated Intensity of the Nucleus”), Average Intensity of the Nuclei (“Average Intensity of the Nucleus”)GFP IMAGES: Integrated Intensity of the Cytoplasm (“Total Cytoplasm Intensity”), Integrated Intensity of the Detected Spots (“Total Spot Intensity”), Ratio of the Integrated Spot to Integrated Cytoplasm Intensities (“Ratio of Spot Intensity to Cytoplasmintensity”), Number of Spots per Cell (“Average Spots Per Cell”)
The primary screen was performed at a compound concentration of 10 µM in 384-well format. The average Z’ for the screen was 0.65 and Z’ values ranged from 0.44 to 0.82 using the ratio of the GFP intensity of the spots over the GFP intensity of the cytoplasm (“Ratio of Spot Intensity to Cytoplasmintensity”) as the primary assay read-out.
Confirmation Assays
Initial hit confirmation of compound solutions resupplied by the MLSMR was done at a single compound concentration (10 µM) in duplicates using the primary screen assay to confirm activity of the hit compounds. Compounds with confirmed activity at 10 µM were tested from stock solutions resupplied by the MLSMR in 7-point dose responses (0.5 to 32 µM) to evaluate potency. Potent compounds (IC50 <5 µM) were clustered into scaffolds and 10-point dose responses (0.06 to 32 µM) were performed for dry powder compounds selected from hits and their commercially available analogs.
Counterscreen/Selectivity Assays
To eliminate artifacts introduced by the β-arrestin-GFP assay technology, an image-based high-content assay using the same assay technology was performed against the putative cannabinoid receptor GPR55 in antagonist mode. In addition to eliminating false positives caused by assay artifacts, this assay also evaluates selectivity of the GPR35 hit compounds against the GPR55 receptor. This is of additional interest since GPR35 and GPR55 share ~30% identity (2–5). In addition, GFP intensity of the cells was quantified to identify compounds causing cellular fluorescence resulting in a decrease in number of detected spots and thus false positive results.
Secondary Probe Characterization Assays
The identified GPR35 antagonist probes are characterized further by two assays performed in the assay provider’s and his collaborator’s labs. This checks for assay technology artifacts using the same assay technology on another receptor, the vasopressin receptor, which has been well characterized and is not related to GPR35. Any compounds that were also active in the vasopressin antagonist assay would be likely interfering with the GFP-tagged β-arrestin and thus identified as false positives. The second assay evaluates ERK1/2 activity downstream in the GPR35 signaling pathway.
iii. Summary of Results
The GPR35 antagonist primary screen of 291,994 compounds resulted in 549 compounds that were considered as hits using the hit criteria of >50% activity as compared to cells without agonist addition, >30 cells in the imaged area of the well, and a total GFP intensity of <10,000,000 relative units. The upper limit for the total GFP intensity was added as hit criterion to eliminate cell-permeable autofluorescent compounds interfering with detection of spot formation.
Stock solutions resupplied by the MLSMR of 490 compounds were tested for hit confirmation. Single point confirmations at 10 µM concentration were conducted in duplicate on these compounds. 102 of these compounds confirmed using the same hit criteria as for the primary screening campaign. Further testing of these compounds using a seven-point dose response (0.5 to 32 µM concentration range) identified 33 compounds with an IC50 of less than 1 µM and 57 compounds with an IC50 between 1 and 10 µM.
The hits were clustered into scaffolds by using a maximum-common-substructure-based algorithm. Analyzing the assay data in terms of scaffolds therefore, yielded 22 hits from 8 scaffolds and 38 of their commercially available analogs were selected for dry powder purchase. Testing of these dry powder compounds in 10-point dose responses using the primary screen assay and the GPR55 antagonist counterscreen/selectivity assay resulted in 26 compounds with IC50 < 5 µM and GPR55 antagonist selectivity of IC50(GPR55) > 10x IC50(GPR35) spanning 5 scaffolds (Figure 1).
Figure 1
The GPR35 Antagonist Probe Flowchart summarizes the compound triage and decision tree for advancement of the compounds.
The critical path for the development of the probes is shown below:
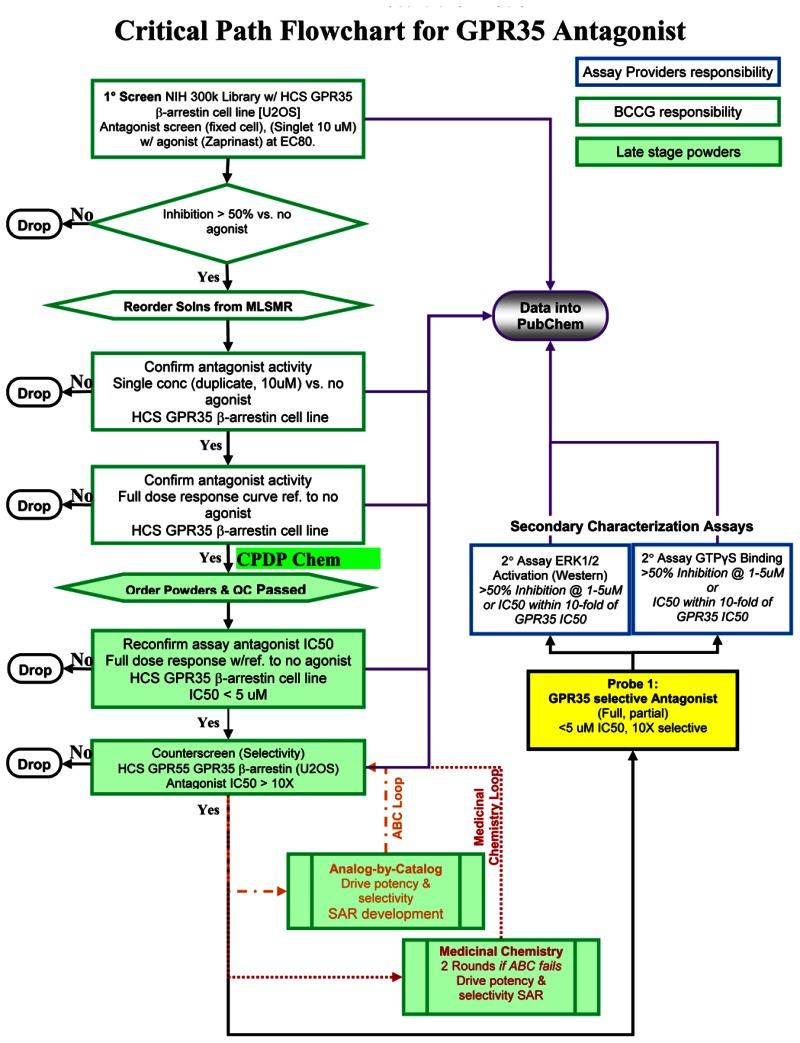
c. Probe Optimization
i. Description of SAR & chemistry strategy (including structure and data) that led to the probe
As shown in Table 3, the probe compound, CID2286812 (SID-87544499) and 5 close analogs all show potent GPR35 antagonism, are selective over GPR55. This class of selective GPR35 antagonists consists of a thioxothiazolidinone with an aromatic or styryl portion consisting of substituent R1 and a 3-carbon spacer connected to a pendant amidobenzoic acid moiety having substitutions R2 and R3. The styryl substituent at R1 provides the most potent and selective inhibitors, as is seen in compounds CID2286812 and CID2286888. While differentially substituted aromatic groups at R1 still provide potent antagonists, the overall profile of the styryl-substituted examples is superior. Substitutions of the amidobenzoic acid portion, R2 and R3, show tolerance for the ortho and meta positions of this ring. The most potent compounds have the carboxylic acid moiety in the para position, with either hydroxyl or H at the meta position. These examples constitute clear SAR of those features required for selective GPR35 antagonism and provide multiple compounds exceeding the set probe criteria.
Table 3
Probe #1: SAR of selective GPR35 antagonists, including probe CID2286812.
The SAR of the second probe CID1542103 and related analogs can be seen in Table 4. This probe series consists of a pyrazolo-pyrimidine core containing a substituted piperazine moiety (Table 4, R1) on the pyrimidine ring and a pendant aromatic group (R2) connected to the pyrazole portion. A variety of aryl groups are tolerated at the R1 position, providing selective GPR35 antagonists in the single-digit micromolar range for GPR35 IC50 and no GPR55 inhibitory activity. Briefly, at R1, ortho substituted groups provide the most potent derivatives as seen with the probe CID1542103 and the close analogue CID1502520. Removal of all substitution from the aryl ring at R1 shows a decrease in GPR35 antagonism, as in CID8056691 and CID8056641. Finally, para substitution here leads to the weakest antagonists, CID661907 and CID2474060. At R2, para substitution is favored, with chloro and methyl substituents providing superior antagonists. It can be noted from CID2474060 that substitution at positions other than the para position result in a loss of GPR35 antagonist activity.
Table 4
Probe #2: SAR of selective GPR35 antagonists, including probe CID1542103.
3. Probe(s)
a. Chemical name of probe compound (s) (Chemical IUPAC)
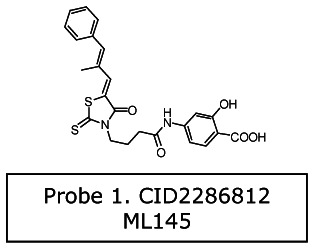
Probe 1 (series 1): 2-hydroxy-4-[4-[(5Z)-5-[(E)-2-methyl-3-phenylprop-2 enylidene]-4-oxo-2- sulfanylidene-1,3-thiazolidin-3-yl]butanoylamino] benzoic acid [ML145]
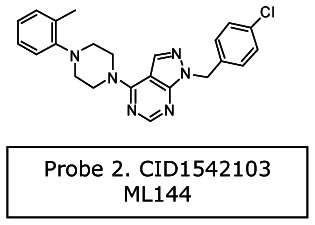
Probe 2 (series 2): 1-[(4-chlorophenyl)methyl]-4-[4-(2-methylphenyl)piperazin-1-yl]pyrazolo[3,4-d]pyrimidine [ML144]
b. Probe chemical structure(s) including stereochemistry
c. Structural Verification Information of probe SID
The probe SIDs are for Probe #1 SID-87544499 (corresponding to CID2286812) and Probe #2 SID-87544496 (corresponding to CID1542103) please see below at the spectra.
(1) PubChem SID-87544499:
NMR Purity
>95% (1H-NMR): 1H NMR (400 MHz, DMSO-d6) δ ppm:
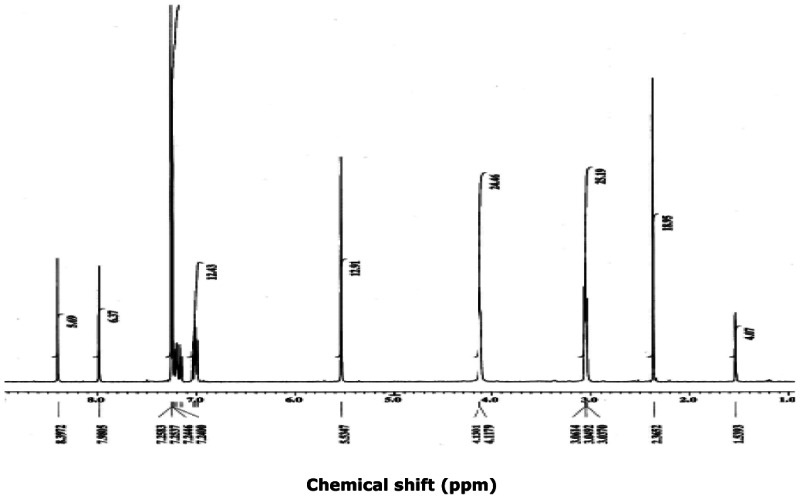
Purity = 98% (HPLC-MS)
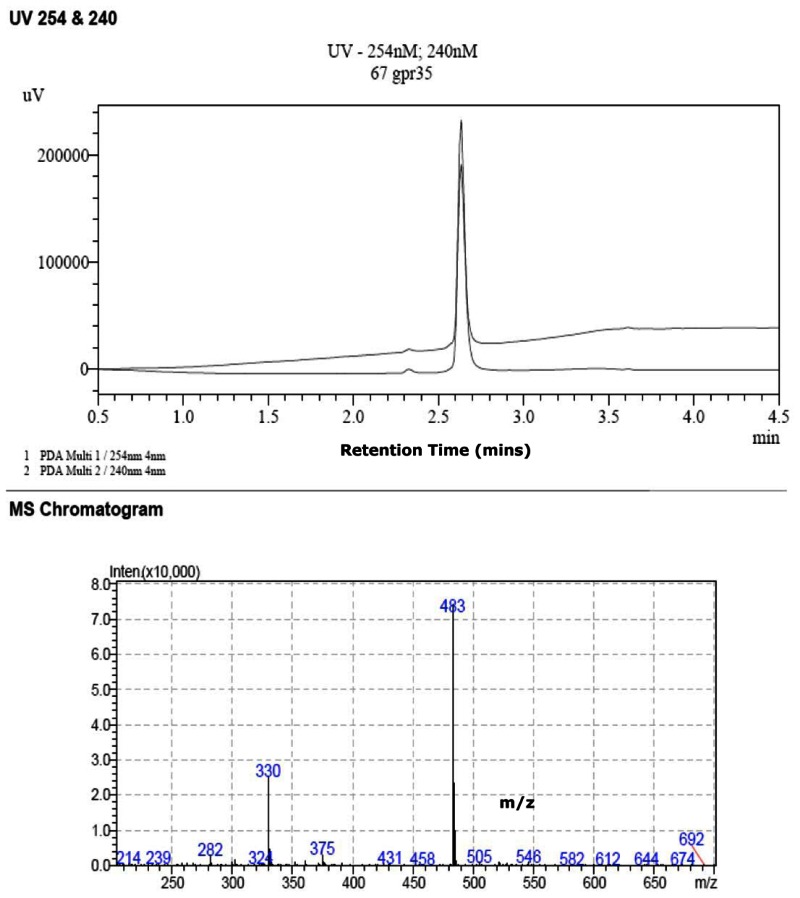
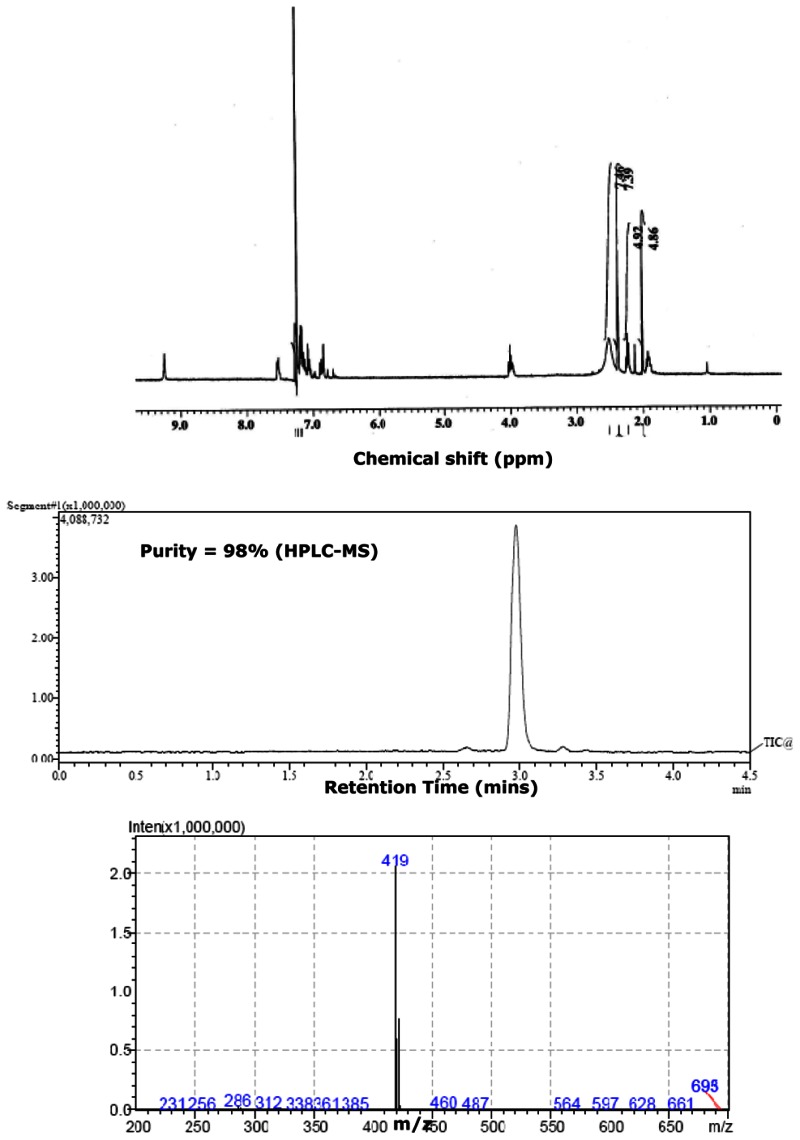
Mass Spec
SID-87544499: ESI-MS m/z calcd for C24H22N2O5S2 [M+H]+: 483, found: 483.
SID-87544496: ESI-MS m/z calcd for C23H23ClN6 [M+H]+: 419, found: 419.
d. PubChem CID(s) (corresponding to the SID)
For Probe #1: PubChem CID-2286812 (corresponding to the SID-87544499).
For Probe #2: PubChem CID-1542103 (corresponding to the SID-87544496).
e. Availability from a vendor
- Scaffold 1, CID2286812 is commercially available from ChemBridge CAT # 6791281
- Scaffold 2, CID1542103 is commercially available ChemBridge: Cat#7652281
f. MLS#'s of probe molecule and five related samples that were submitted to the SMR collection
Table 5A
Submission information on Probe 1: CID2286812 and analogs.
Table 5B
Submission information on Probe#2: CID1542103 and analogs.
g. Mode of action for biological activity of probe
Probes identified in this project are acting at the beginning of the GPR35 signaling pathway. Ligand binding causes phosphorylation of the GPCR, which in turn causes translocation of the β-arrestin to the membrane, where it binds to the GPCR. The β-arrestin-GPCR complex internalizes into clathrin-coated pits within the cell, where it dissociates and the receptor recycles back to the membrane. Since the assay read-out quantifies formation of β-arrestin-GPCR pits, the identified antagonists interfere with the pit formation or any process upstream. The selectivity of these two probes for GPR35 (as described in our Critical Path Flowchart above), but not for the related GPR55 orphan receptor in a cognate β-arrestin HCS assay supports that they are not non-specifically interfering with signaling directly at or downstream of the β-arrestin signaling pathway. In follow-up studies by Dr. Barak (see sec. 5 below), these probes also did not, as expected, agonize nor antagonize β-arrestin mediated signaling of the completely unrelated vasopressin receptor, again supporting their GPR35 specificity. Additional secondary assays by Dr. Abood demonstrated that the probes did inhibit Erk1/2 phosphorylation. This confirms that our imaging assay based results translate to the authentic downstream biological response.
h. Detailed synthetic pathway for synthesizing probe#1
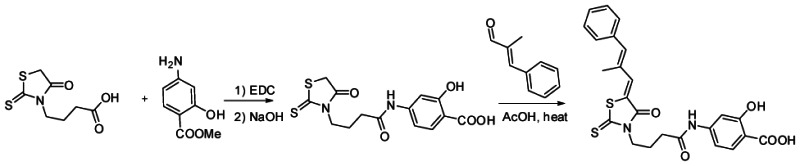
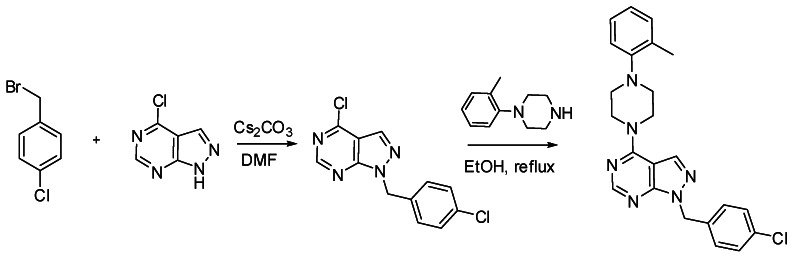
i. Detailed synthetic pathway for synthesizing probe#2
j. Summary of probe properties (solubility, absorbance/fluorescence, reactivity, toxicity, etc.)
In Vitro Pharmacology Profiles of Probes CID2286812 [ML145] and CID1542103 [ML144] (See Table 6 below). Both nominated probes, CID2286812 [ML145] and CID1542103 [ML144] were evaluated in a detailed in vitro pharmacology screen.
Table 6
Summary of in vitro ADMET/PK Properties of the GPR35 Antagonist Probes.
Neither probe molecule had appreciable solubility at pH 5.0. CID2286812 [ML145] had good solubility at the higher pHs (pH 6.2 & pH 7.4), however, the solubility of CID1542103 [ML144] remained poor at the higher pHs.
The PAMPA (Parallel Artificial Membrane Permeability Assay) assay is used as an in vitro model of passive, transcellular permeability. An artificial membrane immobilized on a filter is placed between a donor and acceptor compartment. At the start of the test, drug is introduced in the donor compartment. Following the permeation period, the concentration of drug in the donor and acceptor compartments is measured using UV spectroscopy. In this assay CID2286812 [ML145] has a bell-shaped permeability with good permeability at pH 6.2, while permeability decreased to moderate/poor levels at pH 5 and pH 7.4. In contrast, CID1542103 [ML144] has moderate permeability at pH 5.0, but undetectable or poor permeability at pH 6.2 and 7.4.
Plasma Protein Binding is a measure of a drug’s efficiency to bind to the proteins within blood plasma. The less bound a drug is, the more efficiently it can traverse cell membranes or diffuse. Highly plasma protein bound drugs are confined to the vascular space, thereby having a relatively low volume of distribution. In contrast, drugs that remain largely unbound in plasma are generally available for distribution to other organs and tissues. Both CID2286812 [ML145] and CID1542103 [ML144] are highly bound (98–99.9%) to both human and mouse plasma.
Plasma Stability is a measure of the stability of small molecules and peptides in plasma and is an important parameter, which strongly can influence the in vivo efficacy of a test compound. Drug candidates are exposed in plasma to enzymatic processes (proteinases, esterases), and they can undergo intramolecular rearrangement or bind irreversibly (covalently) to proteins. Interestingly, CID2286812 [ML145] shows some instability in human plasma (77%), but more stable to mouse plasma (94%). In contrast, CID1542103 [ML144] shows excellent stability (90 & 100%) in both human and mouse plasma.
The microsomal stability assay is commonly used to rank compounds according to their metabolic stability. This assay addresses the pharmacologic question of how long the parent compound will remain circulating in plasma within the body. CID2286812 [ML145] shows poor stability (2.4% & 7.0%) in both human and mouse liver homogenates. By comparison CID1542103 [ML144] shows modest hepatic microsome stability (57% & 31%) in both human and mouse liver homogenates.
CID2286812 [ML145] shows no toxicity (>50 µM) toward human hepatocyctes, whereas CID1542103 [ML144] yields some moderate toxicity in our assay.
A recent retrospective study (February 4, 2010) describes a number of substructural features that identifies compounds as “frequent hitters” or promiscuous in a number of biochemical high-throughput screens (11). The authors recommended that Pan Assay Interference Compounds (PAINS) be excluded from bioassay libraries, as they have been shown to be generally interfering of HTS assays through general chemical reactivities, chelation, aggregation, spectroscopic overlaps and have also been cited as “cul de sac” compounds that waste resources. One of the substructures of concern, the rhodanine element (see structure), is present in our first probe molecule, CID2286812, therefore raises some concerns for the potential promiscuosity for this probe. However, this probe has been shown to be strongly potent yet selective against the related GPR55 receptor in the same assay format, so general PAINS reactivity does not appear to be present. Furthermore during the initial scaffold triage, two close analogs of this probe molecule were not active in any of the 70–72 assays in PubChem in which they were tested. From this report, we surmise that rhodanine reactivity is dependent upon having an unsubstituted ring nitrogen or substituents that are in aromatic conjugation with it. In our probe, CID 2286812, this nitrogen has an aliphatic chain substituent that yields a tertiary amine that renders this rhodanine less reactive.
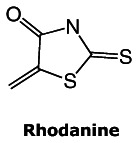
The 2nd probe, CID1542103, does not have these concerning features, and indeed was found to score in only 11 of 354 assays when it was tested in PubChem. While this probe is 100-fold less potent than the 1st probe and not as dramatically pro forma selective against GPR55 compared to CID2286812, it still does meet the probe criteria set by the team and the assay provider. Furthermore, based on its narrow spectrum of cross-reactivity as assessed by its very low hit rate in over 350 PubChem assays, it is expected to be a useful and selective tool compound for studying GPR35.
k. Probe properties
Table 7Properties computed from Structure
| Calculated Property | Probe Identity | |
|---|---|---|
| CID2286812 (scaffold 1) | CID1542103 (Scaffold 2) | |
| MLS-0205231 | MLS-0013034 | |
| Molecular Weight [g/mol] | 482.57188 | 418.92192 |
| Molecular Formula | C24H22N2O5S2 | C23H23ClN6 |
| XLogP3-AA | 5.1 | 4.8 |
| H-Bond Donor | 3 | 0 |
| H-Bond Acceptor | 5 | 5 |
| Rotatable Bond Count | 8 | 4 |
| Tautomer Count | 17 | - |
| Exact Mass | 482.097013 | 418.167272 |
| MonoIsotopic Mass | 482.097013 | 418.167272 |
| Topological Polar Surface Area | 164 | 50.1 |
| Heavy Atom Count | 33 | 30 |
| Formal Charge | 0 | 0 |
| Complexity | 835 | 548 |
| Isotope Atom Count | 0 | 0 |
| Defined Atom StereoCenter Count | 0 | 0 |
| Undefined Atom StereoCenter Count | 0 | 0 |
| Defined Bond StereoCenter Count | 2 | 0 |
| Undefined Bond StereoCenter Count | 0 | 0 |
| Covalently-Bonded Unit Count | 1 | 1 |
4. Comparative data showing probe specificity for target in biologically relevant assays
There is no current state of art probes except from this probe report. Further follow-up work has been conducted in the assay provider’s laboratories (Dr. Barak and collaborator Dr. Abood) as outlined in the CPDP as post Probe Nomination characterization. The three additional downstream assays are as described below and the SAR data summarized for the 1st probe series (Table 8A) and 2nd probe series below (Table 8B).
Table 8A
Comparative Downstream and Selectivity assays by Assay Provider for CID2286812.
Table 8B
Comparative Downstream and Selectivity assays by Assay Provider for CID1542103.
Assay for Agonists (AID-463201) and Antagonists (AID-463202) of Vasopressin Receptor Type IIa [GI: 208973254; Gene: 554]
This assay was originally proposed as the primary selectivity assay, but as our Center was already performing the cognate GPR55 assays for a different project, and since it was an “in family” target, we agreed substitute GPR55 as the selectivity filter in the Critical Path Flowchart (above on p. 7). These assays were performed by the assay provider and characterize the selectivity of compounds against the unrelated vasopressin receptor. It is a confirmatory assay for the final probes and analogs that are already known to be selective against GPR55. This imaging assay utilizes a cell line permanently expressing a beta-arrestin GFP biosensor and human vasopressin receptor type IIa. Upon agonist-mediated GPCR activation, the arrestin-GFP redistributes from the cytosolic compartment to the plasma membrane to coated pits. This arrestin-GFP redistribution is measured as increased local concentrations of fluorescent arrestins. As can be seen in Tables 8A and 8B, both probes and analogs do not agonize (<1% activation) nor antagonize (< 37% inhibition) the vasopressin receptor. We note the high variance in the vasopressin antagonist assays.
Assay for ERK1/2 Activity (AID-463217) in GPR35-Overexpressing U2OS Cells [GI: 33695097; Gene: 2859]
Assay performed by the assay provider characterizes the downstream ERK phosphorylation activity of probe compounds. This is an “in-cell” Western assay utilizes a cell line permanently expressing a beta-arrestin GFP biosensor and human GPR35 receptor. Upon agonist-mediated GPCR activation, ERK1/2 phosphorylation occurs as measured by pERK1/2 antibodies. The ERK1/2 assay is generally much less sensitive than the beta-arrestin GPCR assays and the assay protocol does not allow for compound pre-incubation before agonist addition, which may result in much higher IC50 values (10 – 40-fold) for the pERK-ICW potencies as compared the GPR35 assay. This can be seen in Tables 8A where the nanomolar GPR35 probe and analogs yield micromolar potency in ERK phosphorylation. For the less potent probe and analogs in Table 8B, the rightward shift in potency in ERK limits the obtainment of a good IC50 due to solubility limits. We note that while there are some rank order differences in the apparent pERK IC50’s versus the GPR35 IC50s, possibly due different solubility or ADME properties, the overall conclusion is that both probe scaffold classes do have measureable activity in the downstream assays on the authentic signaling pathway.
5. References
- 1.
- O’Dowd BF, Nguyen T, Marchese A, Cheng R, Lynch KR, Heng HH, Kolakowski LF Jr, George SR. Discovery of three novel G-protein-coupled receptor genes. Genomics. 1998;47:310–3. [PubMed: 9479505]
- 2.
- Guo J, Williams DJ, Puhl HL 3rd, Ikeda SR. Inhibition of N-type calcium channels by activation of GPR35, an orphan receptor, heterologously expressed in rat sympathetic neurons. J Pharmacol Exp Ther. 2008;324:342–51. [PubMed: 17940199]
- 3.
- Johns DG, Behm DJ, Walker DJ, Ao Z, Shapland EM, Daniels DA, Riddick M, Dowell S, Staton PC, Green P, Shabon U, Bao W, Aiyar N, Yue TL, Brown AJ, Morrison AD, Douglas SA. The novel endocannabinoid receptor GPR55 is activated by atypical cannabinoids but does not mediate their vasodilator effects. Br J Pharmacol. 2007;152:825–31. [PMC free article: PMC2190033] [PubMed: 17704827]
- 4.
- Ryberg E, Larsson N, Sjogren S, Hjorth S, Hermansson NO, Leonova J, Elebring T, Nilsson K, Drmota T, Greasley PJ. The orphan receptor GPR55 is a novel cannabinoid receptor. Br J Pharmacol. 2007;152:1092–101. [PMC free article: PMC2095107] [PubMed: 17876302]
- 5.
- Taniguchi Y, Tonai-Kachi H, Shinjo K. Zaprinast, a well-known cyclic guanosine monophosphate-specific phosphodiesterase inhibitor, is an agonist for GPR35. FEBS Lett. 2006;580:5003–8. [PubMed: 16934253]
- 6.
- Brown AJ. Novel cannabinoid receptors. Br J Pharmacol. 2007;152:567–75. [PMC free article: PMC2190013] [PubMed: 17906678]
- 7.
- Ohshiro H, Tonai-Kachi H, Ichikawa K. GPR35 is a functional receptor in rat dorsal root ganglion neurons. Biochem Biophys Res Commun. 2008;365:344–8. [PubMed: 17996730]
- 8.
- Okumura S, Baba H, Kumada T, Nanmoku K, Nakajima H, Nakane Y, Hioki K, Ikenaka K. Cloning of a G-protein-coupled receptor that shows an activity to transform NIH3T3 cells and is expressed in gastric cancer cells. Cancer Sci. 2004;95:131–5. [PubMed: 14965362]
- 9.
- Shrimpton AE, Braddock BR, Thomson LL, Stein CK, Hoo JJ. Molecular delineation of deletions on 2q37.3 in three cases with an Albright hereditary osteodystrophy-like phenotype. Clin Genet. 2004;66:537–44. [PubMed: 15521982]
- 10.
- Barak LS, Ferguson SS, Zhang J, Martenson C, Meyer T, Caron MG. Internal trafficking and surface mobility of a functionally intact beta2-adrenergic receptor-green fluorescent protein conjugate. Mol Pharmacol. 1997;51:177–84. [PubMed: 9203621]
- 11.
- Baell BJ, Holloway AG. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J Med Chem. 2010 2010 Feb 4; [Epub ahead of print] [PubMed - as supplied by publisher] [PubMed: 20131845]
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Selective GPR35 Antagonists - Probe 3.[Probe Reports from the NIH Mol...]Review Selective GPR35 Antagonists - Probe 3.Heynen-Genel S, Dahl R, Shi S, Sauer M, Hariharan S, Sergienko E, Dad S, Chung TDY, Stonich D, Su Y, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review The therapeutic potential of orphan GPCRs, GPR35 and GPR55.[Front Pharmacol. 2015]Review The therapeutic potential of orphan GPCRs, GPR35 and GPR55.Shore DM, Reggio PH. Front Pharmacol. 2015; 6:69. Epub 2015 Apr 15.
- 8-Benzamidochromen-4-one-2-carboxylic acids: potent and selective agonists for the orphan G protein-coupled receptor GPR35.[J Med Chem. 2013]8-Benzamidochromen-4-one-2-carboxylic acids: potent and selective agonists for the orphan G protein-coupled receptor GPR35.Funke M, Thimm D, Schiedel AC, Müller CE. J Med Chem. 2013 Jun 27; 56(12):5182-97. Epub 2013 Jun 17.
- Review GPR55 and GPR35 and their relationship to cannabinoid and lysophospholipid receptors.[Life Sci. 2013]Review GPR55 and GPR35 and their relationship to cannabinoid and lysophospholipid receptors.Zhao P, Abood ME. Life Sci. 2013 Mar 19; 92(8-9):453-7. Epub 2012 Jul 20.
- Review Screening for Selective Ligands for GPR55 - Antagonists.[Probe Reports from the NIH Mol...]Review Screening for Selective Ligands for GPR55 - Antagonists.Heynen-Genel S, Dahl R, Shi S, Milan L, Hariharan S, Sergienko E, Hedrick M, Dad S, Stonich D, Su Y, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Selective GPR35 Antagonists - Probes 1 & 2 - Probe Reports from the NIH Molecula...Selective GPR35 Antagonists - Probes 1 & 2 - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...
