Reproduced from Annu. Rev. Microbiol. 2001. 55:709 – 42.
Key Words: phylogenetic analysis, gene acquisition, gene displacement, adaptation, orthologs
Comparative analysis of bacterial, archaeal, and eukaryotic genomes indicates that a significant fraction of the genes in the prokaryotic genomes have been subject to horizontal transfer. In some cases, the amount and source of horizontal gene transfer can be linked to an organism’s lifestyle. For example, bacterial hyperther-mophiles seem to have exchanged genes with archaea to a greater extent than other bacteria, whereas transfer of certain classes of eukaryotic genes is most common in parasitic and symbiotic bacteria. Horizontal transfer events can be classified into distinct categories of acquisition of new genes, acquisition of paralogs of existing genes, and xenologous gene displacement whereby a gene is displaced by a horizontally transferred ortholog from another lineage (xenolog). Each of these types of horizontal gene transfer is common among prokaryotes, but their relative contributions differ in different lineages. The fixation and long-term persistence of horizontally transferred genes suggests that they confer a selective advantage on the recipient organism. In most cases, the nature of this advantage remains unclear, but detailed examination of several cases of acquisition of eukaryotic genes by bacteria seems to reveal the evolutionary forces involved. Examples include isoleucyl-tRNA synthetases whose acquisition from eukaryotes by several bacteria is linked to antibiotic resistance, ATP/ADP translocases acquired by intracellular parasitic bacteria, Chlamydia and Rickettsia, apparently from plants, and proteases that may be implicated in chlamydial pathogenesis.
Introduction
Horizontal (lateral) gene transfer, the transfer of genes between different species, is an evolutionary phenomenon whose extent and even very existence have been the subject of a longtime debate that tends to become particularly vigorous when cases of horizontal gene transfer that involve eukaryotes are considered (98, 103). This is understandable because (a) horizontal gene transfer seems to challenge the traditional, tree-based view of the evolution of life and the core neo-Darwinist belief in the central role of reproductive isolation between species in evolution (21–23, 53, 74, 89, 90), concepts that, at least initially, have been developed in studies on the evolution of sexually reproducing eukaryotes, and (b) like many evolutionary phenomena, horizontal transfer is hard to prove unambiguously. Until the advent of the genome sequencing era, although striking anecdotal examples of horizontal gene transfer have been described (97) and some prescient speculation on the potential major evolutionary impact of such events has been published (102), the prevailing opinion seemed to be that this phenomenon was rare enough to be insignificant for our general understanding of evolution. The only instance where the importance of horizontal gene transfer had been clearly recognized was the apparent flow of genes from the genomes of endosymbiotic organelles—mitochondria in all eukaryotes and chloroplasts in plants—to the eukaryotic nuclear genome (37, 38, 58, 75).
However, sequenced-based genomics has quickly shown that these “illegitimate” evolutionary events are too common to be dismissed as inconsequential (22, 84). The first strong indication probably came from the multifactorial analysis of codon frequencies in portions of the Escherichia coli genome that indicated significant deviation from the general pattern of codon usage in about 15% of this bacterium’s genes (76). Because some of the genes in this group showed clear relationships with bacteriophage genes, the hypothesis has been proposed that all these genes were alien to E. coli and have been acquired horizontally from various sources. These types of observations seemed to strongly suggest substantial and relatively recent horizontal gene flow. On a different evolutionary scale, the possibility of horizontal transfer being a major factor in evolution has been suggested by the difficulty of constructing congruent phylogenetic trees for different sets of orthologous genes from a wide range of organisms. For example, some archaeal genes showed a clear affinity to their eukaryotic counterparts, whereas others equally strongly clustered with bacterial homologs (12, 33, 40).
The availability of multiple prokaryotic genomes for comparative analysis ushered in the new age of “lateral genomics” (21). Dramatic differences in gene repertoires even among bacteria that belong to the same evolutionary lineage, such as E. coli and Haemophilus influenzae (106), indicated that genome evolution could not be reasonably described in terms of vertical descent alone. It is clear that much of the difference was attributable to differential gene loss, particularly in parasites, but horizontal gene transfer is the other major evolutionary factor that could help explain the emerging complex picture of prokaryotic genomes. The archaeal genomes presented a particularly striking “genomescape” strongly suggestive of massive horizontal gene transfer. In agreement with the earlier indications from phylogenetic studies, but now on the whole-genome scale, it has become clear that archaeal proteins split into those genes that were most similar to their bacterial homologs and that looked “eukaryotic” (24, 56, 70). Some exceptions notwithstanding, the bacterial and eukaryotic proteins in archaea were neatly divided along functional lines, with those involved in information processing (translation, transcription, and replication) showing the eukaryotic affinity, and metabolic enzymes, structural components, and a variety of un-characterized proteins that appeared to be “bacterial.” Because the informational components generally appear to be less subject to horizontal gene transfer (however, some important exceptions are discussed below) and in accord with the standard model of early evolution whereby eukaryotes share a common ancestor with archaea, these observations have been tentatively explained by massive gene exchange between archaea and bacteria (56). This view has been further supported when the genomes of two hyperthermophilic bacteria, Aquifex aeolicus and Thermotoga maritima, were sequenced. Each of these genomes contained a significantly greater fraction of archaeal genes than any of the other bacterial genomes, establishing a plausible connection between the similarity in the lifestyles of evolutionarily distant organisms and the apparent rate of horizontal gene exchange between them (4, 83). Also, these findings emphasized the issue of the adaptive versus opportunistic nature of horizontal gene transfer: Do the genes that might have been acquired from archaea directly enable these bacteria to thrive in hyperthermal conditions, or have they acquired more archaeal genes simply because they have been more exposed to contacts with archaea because of their thermophily? Another case of nonrandomness in the apparent horizontal gene transfer has been observed in the genome of the cyanobacterium Synechocystis sp., which encodes a variety of proteins associated with different forms of signaling that have been thought of as eukaryotic (50, 91).
The finding that the contributions of horizontal gene transfer and lineage-specific gene loss to the gene repertoire of prokaryotes was comparable to that of vertical descent amounted to a major shift in our understanding of evolution. Indeed, it became apparent that, in many cases, phylogenetic trees for different genes were incongruent not because of artifacts inherent in tree-construction methods but because of genuine differences in the evolutionary histories of these genes brought about by horizontal transfer. Thus, a true tree of life, a species tree, could not be constructed, not because of the complexity of the problem and erosion of the phylogenetic signal from ancient divergence events, but perhaps in principle (22, 23). The best one could hope for was a consensus tree that would reflect the history of a gene core conserved in all or the great majority of species and not subject to horizontal gene transfer. But the very existence of such a stable core and more so its actual delineation remain questionable.
In retrospect, the magnitude of apparent horizontal gene transfer in prokaryotes perhaps should not have come as a complete surprise. Indeed, the ability of microbes to absorb DNA from the environment and to integrate it into the genome had been dramatically demonstrated in the Avery-McLeod-McCarthy experiment of 1943 that proved the role of DNA as the genetic material (7). Subsequently, high transformability has been demonstrated for a variety of microbial species (69). Moreover, bacteriophages and plasmids well known to cross-species barriers provide additional, potentially highly effective vehicles for horizontal gene transfer (42, 77, 101). Given the fact that microbes typically coexist in tightly knit communities such as microbial mats and the microflora of animal intestines (49, 79, 93, 104, 111), it appears that opportunities should abound for DNA transfer by various means between diverse prokaryotes and potentially even between eukaryotes and prokaryotes, although in the latter case, the extra complication of getting rid of introns resident in eukaryotic genes is involved.
Despite its obvious growth in prominence with the progress of sequence-based comparative genomics, the issue of horizontal gene transfer as a major evolutionary force remains highly controversial (5, 57, 67). One reason seems to be that the major change in the general picture of evolution seems inevitable if the notion of lateral genomics is vindicated and prompts understandable and perhaps epistemologically justified (the evidence must be fully convincing to justify a paradigm shift) caution in many quarters. The other problem is that, whereas the general significance of horizontal transfer seems to ensue from genome comparisons, the validity of this evolutionary scenario in many individual cases can be questioned owing to uncertainties in phylogenetic tree topologies, unequal evolution rates in different lineages, and other complications.
In this review, we consider the criteria used to ascertain horizontal gene transfer, present conservative quantitative estimates of the amount of likely horizontal gene transfer in the completely sequenced prokaryotic genomes, propose a classification of the distinct types of horizontal transfers, and discuss examples of apparent acquisition of eukaryotic genes by bacteria and archaea.
Criteria for Detecting Horizontally Transferred Genes
All criteria for identifying probable horizontal gene transfer, or more precisely acquisition of foreign genes by a particular genome, inevitably rely on some unusual feature(s) of subsets of genes that distinguishes them from the bulk of genes in the genome. Traditional tests for horizontal gene transfer involve phylogenetic tree analysis and inherit all the pitfalls typical of these methods. However, the accumulation of multiple genome sequences provides for new, perhaps simpler criteria. When considering these criteria, one has to keep in mind that direct proofs for horizontal gene transfer may be unavailable for the simple reason that there is no record of these evolutionary events other than what could be deciphered by comparison of extant genomes. Therefore all indications for horizontal transfer necessarily remain probabilistic, and the point of using different criteria is maximizing the likelihood of these events being identified correctly.
Unexpected Ranking of Sequence Similarity Among Homologs
The suspicion of horizontal gene transfer usually emerges when a gene sequence (or rather a protein sequence because database searches are typically performed at the protein level) from a particular organism shows the strongest similarity to a homolog from a distant taxon. For example, when all protein sequences encoded in a bacterial genome are compared with the entire protein database and the detected (probable) homologs (or hits in the jargon of computational biology) are classified according to their taxonomic origin, a certain fraction of proteins shows the greatest similarity to eukaryotic homologs rather than to those from other bacteria. The size of this fraction depends, evidently, on the genome and also on the cutoff (usually expressed in terms of alignment score or expect value) used to define “more similar” [the taxonomic breakdown of the best hits for all proteins encoded in completely sequenced prokaryotic genomes is available through the genome division of the Entrez retrieval system (108) at http://www.ncbi.nlm.nih.gov:80/PMGifs/Genomes/org.html; then see the “Distribution of BLAST Protein Homologs by Taxa” for individual genomes; see also details below]. These genes make a list of candidates for possible horizontal gene exchange between the given bacterium (or, more precisely, the evolutionary lineage it represents) and eukaryotes. The strength of the claim depends on the cutoff used, but generally the evidence from sequence comparisons should be considered preliminary. To make the case for horizontal transfer convincing, phylogenetic analysis is required.
Unexpected Phylogenetic Tree Topology
Analysis of phylogenetic tree topologies is traditionally the principal means to decipher evolutionary scenarios, including horizontal transfer events (103). Indeed, if, for example, in a well-supported tree, a bacterial protein groups with its eukaryotic homologs to the exclusion of homologs from other bacteria and, best of all, shows a reliable affinity with a particular eukaryotic lineage, the conclusion that horizontal gene transfer is at play seems inevitable. Moreover, in a convincing case like this, even the most likely direction of transfer, from eukaryotes to bacteria, seems clear. It is unfortunate, however, that phylogenetic analysis does not offer such clear-cut solutions in all suspected cases of horizontal gene transfer, not necessarily even in a majority of these cases. It is common knowledge that phylogenetic methods are prone to a variety of artifacts, perhaps the most notorious being long-branch attraction (78). This phenomenon is particularly relevant for the analysis of probable horizontal gene transfer because these events may be accompanied by accelerated evolution, hence long branches in phylogenetic trees. Tree topology is a good indicator of the probable course of evolution only in cases when the critical nodes are strongly supported statistically, by bootstrap analysis or other methods (11, 26). However, many gene (protein) families seem to have undergone “star evolution,” with short internal branches. In such cases, the actual tree topology remains uncertain and phylogenetic analysis becomes useless for verifying the candidate–horizontal transfer events. On a more practical note, phylogenetic analysis is time and labor consuming, critically depends on correct sequence alignments, and is hard to automate without compromising the quality.
Unusual Phyletic Patterns
With many complete genome sequences available, new and relatively simple, but potentially powerful, approaches to evolutionary analysis become feasible. With the systematic delineation of families of orthologs (direct evolutionary counterparts related by vertcial descent), the notion of a phyletic (phylogenetic) pattern has been introduced (29, 30, 105). In the most straighforward formulation, a phyletic pattern is simply the pattern of species present or missing in the given cluster of orthologs. The striking observation made during the construction of the collection of clusters of orthologous groups (COGs) of proteins is the extreme diversity of the observed phyletic patterns (105, 107), most of which include only a small number of genomes (Figure 1). This distribution of COGs by the number of represented species immediately suggests major roles of lineage-specific gene loss and horizontal gene transfer in evolution. Certain types of phyletic patterns, however, appear to signal horizontal transfer in a more specific fashion (Table 1). For example, when a robustly defined set of orthologs shows the presence of a typical “archaeal-eukaryotic” protein in a single bacterial lineage, the odds for the horizontal transfer explanation seem to be high. The B-family DNA polymerase (E. coli DNA polymerase II) is a straightforward example of such obvious horizontal gene transfer. The DNA polymerase is a highly conserved protein, and in particular, the similarity between the gamma-proteobacterial Pol II and its archaeal and eukaryotic homologs is highly significant, which seems to render practically irrelevant the typical objections that other bacteria might in fact encode orthologs that have diverged beyond recognition. The presence of the Pol II gene in several sequenced gamma-proteobacterial genomes seems to rule out any possibility of artifacts such as contamination of a bacterial genome with eukaryotic sequences. In this particular case, there even seems to be an indication as to possible vehicle of gene transfer because family B polymerases are encoded by numerous bacteriophages and animal viruses (9, 51).
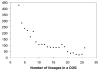
Figure 1
Distribution of the clusters of orthologous groups of proteins (COGs) by the number of represented species. Each COG includes predicted orthologs from at least three genomes that belong to 26 distinct lineages (107).
Table 1
Examples of probable horizontal gene transfers identified using phyletic patterns in COGs a.
Conservation of Gene Order Between Distant Taxa: Horizontal Transfer of Operons
The evolution of bacterial and archaeal genomes involved extensive gene shuffling, and there is little conservation of gene order between distantly related genomes (18, 47, 81). It has been determined that the presence of three or more genes in the same order in distant genomes is extremely unlikely unless these genes form an operon (118). The same analysis implies that each operon typically emerges only once during evolution and is maintained by selection ever after (59, 60). Therefore, when a (predicted) operon is present in only a few distantly related genomes, horizontal gene transfer seems to be the most likely scenario. If such cases can be confirmed by phylogenetic tree analysis for multiple genes comprising the operon, they figure among the strongest indications of horizontal transfer. Horizontal mobility of operons that encode restriction-modification systems is probably the most compelling example of horizontal mobility of operons (52, 82), but for many normal operons, dissemination by horizontal transfer also appears extremely probable; some examples are given in Table 2. The archaeal-type H+-ATPase operon is a well-characterized example of such apparent horizontal dissemination of an operon among bacteria, with displacement of the classical bacterial ATPase operon (43, 85).
Table 2
Examples of horizontally transferred operons.
Anomalous Nucleotide Composition
Anomalous nucleotide composition is widely used but is applicable only to recent horizontal transfers. This approach is based on the “genome hypothesis,” according to which codon usage and GC content are distinct signatures of each genome (35, 36). Thus, genes whose nucleotide or codon composition are significantly different from the mean for a given genome are considered as probable horizontal acquisitions although the likely source of these alien genes generally cannot be identified (31, 61, 76, 80, 84). A significant fraction of prokaryotic genomes, up to 15%–20% of the genes, belongs to this class of recent horizontal acquisitions (31, 61, 76). Many of the horizontally transferred genes revealed by these criteria are prophages, transposons, and other genetic elements for which such evolutionary mobility is not unexpected. This type of horizontal gene transfer and approaches used for its identification have been recently discussed in some detail (84) and are not specifically considered here.
Establishing the Direction of Horizontal Gene Transfer
Difficult as it might be to prove beyond reasonable doubt that horizontal gene transfer has occurred during the evolution of a particular gene family, it is even harder to unequivocally determine which organism is the donor and which one is the recipient in each case. To begin with, the available collection of genome sequences is but a tiny sampling of the genome universe (86), because of which we cannot reasonably hope to identify the true source of any gene present in a given genome. At best, it might be possible to propose a credible hypothesis as to the donor lineage. The logic used to formulate such hypotheses is based primarily on the “out of Africa” principle (15)—which assumes that if horizontal transfer has indeed occurred, the taxon with the most diverse representation of the given family is the most likely source. The examples in Table 1 were selected to illustrate this approach; these gene families are either widely (usually universally) represented in archaea and eukaryotes but are found in only one or a few bacterial species, or vice versa, which strongly suggests the transfer polarity. However, in the rather common cases of a limited representation of a given family in two distant taxa, horizontal transfer per se may be (almost) indisputable, but the direction cannot be established with any confidence.
Classification and Quantification of Horizontal Gene Transfer Events
Horizontal gene transfer events can be classified into at least three distinct categories with respect to the relationships between the horizontally acquired gene and homologous genes (if any) preexisting in the recipient lineage: (a) acquisition of a new gene missing in other members of a given clade, (b) acquisition of a paralog of the given gene with a distinct evolutionary ancestry, and (c) acquisition of a phylogenetically distant ortholog followed by xenologous gene displacement—that is, elimination of the ancestral gene [xenology has been defined as homology of genes incongruent with the species tree and so implies horizontal gene transfer (32, 88)].
In terms of the actual evolutionary scenarios, the first two classes of events in some cases may reflect nonorthologous gene displacement, that is, acquisition of an unrelated (or distantly related) gene with the same function as an essential ancestral gene typical of the given clade, with subsequent elimination of the latter (54, 55).
We sought to quantitatively assess the amount of horizontal gene transfer in bacterial and archaeal genomes and to classify the transfer events, at least tentatively, into the above categories. With all the caveats discussed above, taxonomic classification of database hits is the only practicable method to identify candidate horizontal gene transfer events on a genome scale. Therefore we applied this approach with conservative cutoffs and identified proteins that are significantly more similar to homologs from other taxa than to those from the taxon to which the given species belongs (hereinafter paradoxical best hits and reference taxa). Protein sets from 31 complete prokaryotic genomes (9 archaeal and 22 bacterial) available at the time of this analysis were extracted from the genome division of the Entrez retrieval system (108) and used as queries to search the nonredundant (NR) protein sequence database at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD) with the gapped BLASTP program (1). From the results of these searches, three sets of paradoxical best hits were identified using the Tax Collector program of the SEALS package (110). The first set was designed to include candidate horizontal transfers between phylogenetically most distant organisms. Specifically, for nine archaeal species, all proteins were detected whose best hits to bacterial or eukaryotic proteins expect (E)-values were significantly lower than the E-value of the best hit to an archaeal protein (see Table 3). Similarly, for 22 bacterial proteomes, the paradoxical best hits to archaeal and eukaryotic proteins were collected. A separate subset of the paradoxical best hits was formed by proteins with a significant hit (E <0.001) detected only outside the reference taxon.
Table 3
Candidate horizontal transfers between bacteria, archaea, and eukaryotes: a quantitative assessment a.
The second group of paradoxical best hits was to include candidate gene exchange events between major bacterial lineages. With the current state of genome sequencing, this type of analysis is best applicable to small genomes of parasitic bacteria when at least one larger genome sequence of a related species is available. Thus, for H. influenzae and Rickettsia prowazekii, hits to Proteobacteria (two large proteobacterial genomes, those of E. coli and Pseudomonas aeruginosa, have been sequenced) were compared with hits to all other bacteria, and paradoxical hits were collected. To isolate likely horizontal transfer events between distantly related bacteria, the paradoxical hits to archaea and eukaryotes included in the first set were subtracted. Similarly, for Mycoplasma genitalium, M. pneumoniae, and Ureaplasma urealyticum, the reference taxon was Firmicutes (gram-positive bacteria; available large genomes—Bacillus subtilis and Mycobacterium tuberculosis). In the case of the spirochetes Treponema pallidum and Borrelia burgdorferi, the reference taxon was Spirochaetales (no large genome available for this lineage).
The third set was to include paradoxical best hits owing to probable recent horizontal transfers. For this purpose, two closely related species pairs, namely Chlamydophyla pneumoniae/Chlamydia trachomatis and Mycoplasma genitalium/Mycoplasma pneumoniae were compared. The criteria used to register a paradoxical best hit were the same for all three sets.
Because paradoxical best hits, even with a conservative threshold used in this analysis, provide only a first-approximation estimate of horizontal gene transfer events, a detailed analysis of all candidates was performed for four selected genomes, the bacteria H. influenzae, Vibrio cholerae, and A. aeolicus, and the archaeon Methanobacterium thermoautotrophicum. All paradoxical best hits were examined case by case, which involved establishing the phyletic distribution of the corresponding protein family and constructing phylogenetic trees. Multiple protein sequence alignments were constructed with the ClustalW program (109), checked for the conservation of salient sequence motifs, and used for constructing phylogenetic trees, with the neighbor-joining method (94) as implemented in the NEIGHBOR program of the PHYLIP program package (28).
Table 3 shows the tally of candidate interdomain horizontal transfers for all prokaryotic genomes. In most free-living bacteria, the interdomain transfers seemed to involve ~3% of the genes. This fraction was significantly lower in parasitic bacteria, with the exception of Chlamydia and Rickettsia. In contrast, archaea had a greater fraction of candidate gene transfers of this type, typically between 4% and 8%. It should be emphasized that the protocol used to obtain these numbers will detect primarily relatively recent horizontal transfer events because ancient ones (e.g., those that could have occurred prior to the divergence of the analyzed archaeal species) would have been obscured by interarchaeal hits. For the same reason, the fraction of transfers recorded for each of the gamma-proteobacterial species is likely to be an underestimate given that this lineage is represented by several genomes, including two large ones, E. coli and P. aeruginosa. Given these limitations in detection of paradoxical best hits and the conservative cut-off values used, the level of likely gene exchange between different domains of life seems to be quite substantial.
Several organisms clearly stood out in terms of the number of genes that probably have been horizontally acquired from a different domain of life. In two of these, Synechocystis sp. [a cyanobacterium, the progenitor of chloroplasts (38)] and R. prowazekii [an alpha-Proteobacterium, the group of bacteria to which the progenitor of the mitochondria is thought to belong (2)], the reported high numbers probably reflect the most obvious interdomain horizontal gene transfers, namely those between chloroplasts and mitochondria and eukaryotic nuclear genomes. As noticed previously, hyperthermophilic bacteria, A. aeolicus and especially Thermotoga maritima, are significantly enriched in genes apparently horizontally transferred from archaea (4, 83, 119). Conversely, the archaea Thermoplasma acidophilum and especially Halobacterium sp. appear to possess a much greater number of acquired bacterial genes than other archaeal species, perhaps because these organisms that are moderate thermophiles share their habitats with multiple bacterial species. Along the same line, the difference in the number of acquired genes between two close bacterial species, Bacillus subtilis and B. halodurans, is notable. The excess of archaeal genes in the latter may be plausibly explained by coexistence with halophilic archaea.
In this genome-wide analysis, apparent acquisition of new genes (cases when a given protein simply has no detectable homologs in the reference taxon) could be automatically distinguished from paralog acquisition/xenologous displacement, but differentiating between the latter was not possible without additional detailed study. Notably, the number of probable events of paralog acquisition/xenologous displacement was generally comparable to that of the acquisition of new genes (Table 3).
The data in Table 4 provide a more complete estimate of probable horizontal gene transfer events by using the respective bacterial taxa as the reference taxa for each bacterial genome and accordingly including gene exchange between major lineages. These estimates are expected to be particularly reliable for small genomes because, with the exception of the spirochetes, larger genomes of related bacteria are available, making it unlikely that the paradoxical best hits are due to an insufficient representation of the given taxon in the sequence database. The estimates of horizontal transfer rate obtained by this approach widely differ, from the modest 1.6% of the genes for M. genitalium to the striking 32.6% in T. pallidum. The spirochete data could be an overestimate owing to differential gene loss in the two parasitic spirochetes with different lifestyles (100), but in general, a substantial amount of relatively recent horizontal gene transfer seems to be supported by the data. We also estimated the number of more recent horizontal transfer events by collecting the paradoxical best hits for two pairs of closely related bacterial species—M. genitalium/M. pneumoniae and C. trachomatis/C. pneumoniae. Predictably, this analysis revealed a smaller number of candidate horizontal transfers, with none at all seen in M. genitalium (Table 4).
Table 4
Candidate horizontal gene transfers between major bacterial lineages: a quantitative assessment a.
In an attempt to distinguish between xenologous gene displacement and acquisition of a paralog of a preexisting gene, we performed a phylogenetic analysis of the candidate horizontally transferred genes for three bacterial and one archaeal genomes. In many cases, the phylogenetic tree topology was too complex to make the distinction, but multiple clear-cut cases of both types were identified in each of the analyzed genomes (Table 5 and data not shown). The relative contributions of xenologous gene displacement and paralog acquisition appeared to differ significantly for the compared genomes, with the former phenomenon being prevalent in the hyperthermophilic bacterium A. aeolicus and the archaeon Methanobacterium thermoautotrophicum and the latter in the parasitic bacterium V. cholerae (Table 5).
Table 5
Classification of candidate horizontal gene transfer events in selected genomes a.
The trees in Figure 2 illustrate acquisition of paralogs and xenologous gene displacement. The 3-isopropylmalate dehydrogenase tree (Figure 2A) exemplifies the apparent massive gene exchange between archaea and bacteria, particularly thermophilic ones. Xenologous displacement of the original bacterial gene by the archaeal counterpart seems to have independently occurred in Aquifex, Thermotoga, and Deinococcus. In addition, Thermotoga apparently acquired a paralog from a distinct archaeal source. The 6-phosphogluconate dehydrogenase tree (Figure 2B) shows probable xenologous gene displacement within two pairs of relatively close bacterial species, namely H. influenzae/E. coli and T. pallidum/B. burgdorferi. In the former case, the evolutionary scenario seems clear because the H. influenzae protein belongs to a tight cluster of proteobacterial homologs, whereas the E. coli protein unexpectedly falls within the gram-positive lineage. Therefore the displacement event can be confidently mapped to the E. coli genome. In contrast, it is hardly possible to determine which of the spirochetes has undergone displacement because no orthologous sequences from other species from the same taxon are currently available.
Horizontal Gene Transfer from Eukaryotes to Bacteria and Archaea
Acquisition of eukaryotic genes by bacteria is potentially of particular interest because of the possible role of such horizontally transferred genes in bacterial pathogenicity (39, 84). The amount of such horizontal transfer evaluated using the paradoxical best hit approach seems to be relatively small, typically on the order of 1% of the genes in a given prokaryotic genome (Table 6; the high number of “plant” genes in Synechocystis is an artifact due to the relationship between Cyanobacteria and chloroplasts). There seems to be a modest but consistent excess of acquired eukaryotic genes in at least some parasites, such as M. tuberculosis, P. aeruginosa, Xylella fastidiosa, and C. pneumoniae (Table 1). However, for other parasitic bacteria, such as spirochetes, only a small number of probable acquired eukaryotic genes are detected with this approach. The obvious Cyanobacterium-chloroplast relationship notwithstanding, Synechocystis possesses an unusual excess of eukaryotic genes, including those that are otherwise animal specific, such as proteins that share conserved domains with cadherins and other animal extracellular receptors (91). This could suggest that, similar to the extant cyanobacterial symbionts of poriferans, the ancestors of Synechocystis have passed through a symbiotic phase (87). It is interesting that some apparent acquisition of eukaryotic genes is seen in each of the archaeal genomes, with the greatest number detected in Halobacterium sp. (the archaeal species that also appears to have acquired the greatest number of bacterial genes; see Table 3 and discussion above). A more detailed phyletic breakdown of the eukaryotic acquisitions shows some limited correlations with the parasite-host affinities. For example, there is an apparent excess of animal genes in P. aeruginosa, and an excess of “plant” genes in the plant pathogen X. fastidiosa (Table 6). A paradoxical situation was observed in Chlamydia that seem to have acquired a greater number of genes from plants than from animals. As discussed previously (14), it seems possible that Chlamydiae and their close relatives had a long history of parasitic or symbiotic relationships with eukaryotes and at some stages of their evolution could have been parasites of plants or their relatives.
Table 6
Apparent phylogenetic affinities of eukaryotic best hits in bacteria and archaea.
It should be emphasized that, for the transfer of eukaryotic genes to prokaryotes, the numbers produced in this fashion are likely to be underestimates, perhaps significant, because some of the proteins encoded by the transferred genes may not show highly significant similarity to their eukaryotic ancestors and therefore may be easily missed. This is the case for most signaling proteins discussed below. Apparent acquisition of eukaryotic genes is particularly characteristic of certain functional classes of bacterial genes. Here we discuss two such groups of prokaryotic genes and some notable sporadic cases of apparent horizontal transfer.
Signal Transduction Systems Eukaryotes possess incomparably more versatile signal transduction systems than most bacteria and archaea (19, 45, 62), although in some bacteria, such as Cyanobacteria, Myxobacteria, and Actinomycetes, these systems also show remarkable complexity (25, 44). A detailed survey of the phyletic distribution of eukaryotic signaling domains has shown, rather unexpectedly, their frequent presence (sometimes in highly divergent forms) in prokaryotes (91). Based upon their prevalence among bacteria and archaea, these domains have been classified into those that probably have been inherited from the last universal common ancestor, and those that have evolved in eukaryotes and have been subsequently horizontally transferred to bacteria (and less frequently to archaea). A sampling of the signal transduction domains that show evolutionary mobility and their typical protein contexts in eukaryotes and prokaryotes is presented in Table 7. In cases when a domain is present and involved in an essential function in all eukaryotes and, in contrast, is found in only one or two bacterial lineages, the case for horizontal transfer appears compelling. The SWIB domain, which is present in subunits of the SWI/SNF chromatin-associated proteins in all eukaryotes (14), was found in only one bacterial lineage, Chlamydia (99). In this case, the possibility exists that SWIB domain, one copy of which is fused to the chlamydial topoisomerase I, participates in chromatin condensation, a distinguishing feature of this group of intracellular parasites (8). Similarly, the SET domain, a signature eukaryotic histone methylase (92), has been detected in Chlamydia (99) and X. fastidiosa (95). The role of the SET methylase in these bacteria remains unclear and may point to regulatory mechanisms so far unsuspected in prokaryotes.
Table 7
Examples of eukaryotic-bacterial transfer of genes coding for proteins and domains involved in signal transduction a.
Some of the eukaryotic signaling domains acquired by bacteria probably perform functions that, at least mechanistically, are similar to their functions in eukaryotic systems. This probable functional conservation is exemplified by the phosphoserine–peptide-binding FHA domain (64) whose partners in signal transduction, protein kinases, and phosphatases are represented in both eukaryotes and prokaryotes, indicating a role for this domain in similar phosphorylation-based signaling pathways (63).
Other eukaryotic signaling domains have probably been exapted (34) for completely different functions in bacteria. The examples of such exaptation are the predicted cysteine proteases of two distinct families detected in Chlamydia (71, 99; Table 7). In eukaryotes the adenovirus-type protease family is known (65, 66), and the other family has been predicted (71), to participate in the ubiquitin system of controlled protein degradation. However, the ubiquitin pathway does not exist in bacteria, which rules out functional conservation for these proteins. It is most likely that in Chlamydia, the proteases contribute to the pathogen host cell interaction as indicated, in particular, by the predicted membrane localization of the adenovirus-family proteases in C. trachomatis (99).
Some of the eukaryotic signal transduction domains detected in prokaryotes illustrate the interesting phenomenon of interkingdom domain fusion whereby the acquired eukaryotic domains joined with preexisting bacterial domains within the same protein (117). The aforementioned SWIB domain in Chlamydia is a clear-cut case of fusion of a signaling domain acquired from eukaryotes with a typical bacterial enzyme. The presence of two versions of this domain in the bacterial genome, one that stands alone and one fused to topoisomerase, suggests the probable two-step evolutionary scenario for the origin of such fusions, which includes transfer of a eukaryotic gene followed by recombination with the respective bacterial gene. For reasons that remain unclear, fusion of eukaryotic domains with bacterial ones is particularly common in Actinomycetes (117).
Aminoacyl-tRNA Synthetases
Generally, genes coding for the components of the translation machinery appear to belong to the conserved core of the genome that is less prone to horizontal transfer than are other categories of genes (48). An interesting exception is ribosomal protein S14 for which several probable horizontal transfer events have been revealed by phylogenetic analysis (10). However, aminoacyl-tRNA synthetases (aaRSs) are essential components of the translation machinery whose evolution involves horizontal gene transfer as a common trend that could reflect the relative functional autonomy of these enzymes as opposed, for example, to ribosomal proteins that function as subunits of a tight complex (20, 114, 115). Phylogenetic analysis of the aaRS of all 20 specificities indicated that horizontal transfers have probably occurred in almost each case (115). Several of these events involve transfer of an eukaryotic aaRS to bacteria, typically with replacement of the original bacterial gene (Table 8). Acquisition of eukaryotic aaRS genes is most prominent in two groups of parasitic bacteria, Spirochetes and Chlamydia. That parasites have acquired more aaRS genes than free-living bacteria may not be unexpected, but the reasons why this number is unusually high in spirochetes remain unclear. One plausible possibility is that spirochetes have been identified as parasites or symbionts in a wide variety of eukaryotes (73) and might have had a longer history of such relationships than other groups of parasitic and symbiotic bacteria for which complete genome sequences are currently available. However, spirochetes do not seem to show a general preponderance of horizontal gene transfer from eukaryotes compared with other bacteria (Table 6), which suggests that extensive acquisition of aaRS by these organisms could involve some specific, not yet understood features of their biology.
Table 8
Horizontal transfer of eukaryotic aaRS genes into different bacterial lineages a.
A most unexpected case of probable horizontal transfer from eukaryotes is the TrpRS from the archaeal genus Pyrococcus, which includes free-living, hyperther-mophilic microbes (115). Given this lifestyle, the presence of a eukaryotic aaRS in the Pyrococci (and so far not in any other hyperthermophilic archaea) is almost too unusual to believe, but the result of phylogenetic analysis in this case has been unequivocal. Thus, it seems most likely that Pyrococcus indeed has acquired a eukaryotic TrpRS gene from a thermophilic eukaryote such as a polychaete annelid (96).
In all cases, with one notable exception discussed below, the apparent horizontal transfer of eukaryotic aaRS genes into bacteria involves xenologous gene displacement, i.e., the corresponding ancestral bacterial (archaeal) aaRS is never present along with the eukaryotic one. GlnRS that apparently first emerged in eukaryotes through a duplication of the GluRS gene, and subsequently has been horizontally acquired by gamma-Proteobacteria, is a striking case of nonorthol-ogous gene displacement. In most other bacteria, glutamine incorporation into protein is mediated by a completely different mechanism, namely transamidation, whereby glutamine is formed from Glu-tRNAGln in a reaction catalyzed by the specific transamidation complex GatABC (16, 46). Thus, in this case, horizontal gene transfer accompanied by nonorthologous gene displacement has resulted in a switch to a completely different pathway of an essential biochemical process. In several Proteobacteria, for which only partial genome sequences are available, and in Deinococcus radiodurans, GlnRS and the GatABC complex coexist (41), indicating that in gamma-Proteobacteria the loss of the transamidation system has been a relatively late event compared with the acquisition of the eukaryotic GlnRS gene. The Deinococcus system has been studied in detail, and in this organism, the transamidation mechanism only functions to produce Asn-tRNAAsn from mischarged Asp-tRNAAsn, whereas the formation of Gln-tRNAGln is mediated exclusively by GlnRS (17). Notably, the Deinococcus GlnRS contains a fused domain homologous to the C-terminal domain of the tRNA-recognizing GatB protein, which is thought to increase the specificity toward tRNAGln (72). This case illustrates the intricate functional complexity that may result from gene acquisition by horizontal gene transfer accompanied by interkingdom domain fusion.
It has been proposed that the topology of some of the aaRS trees, particularly that for IleRS, could be accounted for by postulating just one horizontal gene transfer from eukaryotes, with subsequent dissemination among bacteria (13). This is a plausible hypothesis that is compatible with the reliable clustering of all bacterial species that are suspected to have acquired the respective eukaryotic gene in the IleRS and the HisRS trees. However, the topologies of the trees for MetRS, ArgRS, and Asp-AsnRS are not compatible with the single-transfer scenario, which instead suggests multiple cases of acquisition of the respective eukaryotic genes by different bacterial lineages (115).
The gene for the eukaryotic-type IleRS disseminates through bacterial populations on plasmids, conferring resistance to the antibiotic mupirocin (13). This is one of the rare cases when not only the vehicle of horizontal transfer appears clear, but the nature of the selective pressure that results in the fixation of the transferred gene in the bacterial population seems obvious.
Miscellaneous Eukaryotic Genes Acquired by Prokaryotes
Numerous genes that appear to have been horizontally transferred from eukaryotes to bacteria or archaea cannot be conveniently classified into just one or a few functional categories; examples of such diverse genes are given in Table 9. It should be admitted that, on most occasions, the selective advantage that could be conferred on the prokaryote by the acquired eukaryotic gene cannot be easily gleaned from comparative sequence analysis. Nevertheless, the few exceptions when this is feasible allow biologically interesting inferences. Perhaps the most clear-cut example is the chloroplast-type ATP/ADP translocase detected in Chlamydia, Rickettsia, and the plant pathogen X. fastidiosa. The advantage of having this enzyme for the intracellular parasites Chlamydia and Rickettsia is obvious because it allows them to scavenge ATP from the host, thus becoming, at least in part, “energy parasites” (113). However, the discovery of the ATP/ADP translocase in X. fastidiosa (M.Y. Galperin, V. Anantharaman, L. Aravind & E.V. Koonin, unpublished data) is unexpected and might indicate that such use of the energetic facilities of the host is, after all, not limited to bacteria that grow inside host cells. The acquisition of the ATP/ADP translocase by bacteria from plants is beyond reasonable doubt. Furthermore, it seems that this has occurred on at least two independent occasions. Unexpectedly, phylogenetic analysis of bacterial and plant translocases indicated that Chlamydia and Rickettsia probably have exchanged these genes (116). The most plausible hypothesis has it that the gene was first acquired from plants by ancestors of Chlamydia that might have been plant parasites (see above) and was subsequently passed to Rickettsia.
Table 9
Examples of eukaryotic-prokaryotic transfer of functionally diverse genes.
Another case when the adaptive value of the acquired gene seems clear is the sodium/phosphate cotransporter that was detected in V. cholerae but not in any other bacterium; this transporter probably facilitates the survival of the bacterium in the host gastrointestinal tract and could be directly relevant for pathogenesis. A variety of enzymes acquired by bacteria from eukaryotes appear to have been exapted for various functions including interactions with their eukaryotic hosts. One probable example of this is a hemoglobinase-like protease that is encoded by the genome of Pseudomonas (PA4016) and that functions as a virulence factor that degrades host proteins. The fukutin-like enzymes found in certain pathogenic bacteria, such as Haemophilus and Streptococcus, represent another case of horizontally transferred eukaryotic proteins apparently adapted by these bacteria for the modification of their own surface molecules (3). In some cases, such as an α/β-fold hydrolase [either a lipolytic cutinase or a related esterase (68)] that has probably been acquired by Mycobacterium from fungi, the horizontally transferred gene has apparently undergone functional diversification on entry into the bacterial lineage through a series of lineage-specific gene duplications.
On a more general note, the acquisition and utilization of eukaryotic genes by prokaryotes shows the remarkable functional plasticity of many cellular systems that is manifested in the compatibility of components evolved in phylogenetically distant organisms. A striking example is topoisomerase IB, an enzyme that is ubiquitous in eukaryotes but that had not been seen in the prokaryotic world until the genome of the extreme radioresistant bacterium D. radiodurans had been sequenced (112). Despite the major differences in the protein composition and mechanisms between the bacterial and eukaryotic repair systems (6, 27), this eukaryotic topoisomerase contributes to the ultraviolet resistance of the bacterium and hence apparently does have a function in repair (72).
Conclusions
Rough estimates based upon the analysis taxon-specific best hits indicate a high level of horizontal gene transfer for most bacterial and archaeal genomes. For selected genomes analyzed in detail, this is largely confirmed by phylogenetic analysis. Probable horizontal transfer events could be classified into the distinct categories of acquisition of new genes and acquisition of paralogs, sometimes followed by nonorthologous gene, and acquisition of phylogenetically distant orthologs followed by xenologous gene displacement. The rates of these different types of horizontal gene transfer events seem to be generally comparable although their relative contributions differ among bacterial and archaeal lineages.
Acquisition of eukaryotic genes by bacterial genomes, particularly those of parasites, and symbionts, and, to a lesser extent, by archaeal genomes, is one of the important directions of horizontal gene flow. Apparent horizontal gene transfer was detected in various functional classes of genes, although it is particularly characteristic of certain categories, such as aminoacyl-tRNA synthetases and different signal transduction systems.
From an evolutionary-theoretical prospective, horizontal transfer, particularly when it occurs between eukaryotes and bacteria, is a testimony to the remarkable unity of molecular-biological mechanisms in all types of cells that result in the compatibility of eukaryotic and bacterial proteins that have evolved in their distinct milieux for billions of years. Although coadaptation of proteins in the course of evolution might impede horizontal transfer of certain types of genes (e.g., those coding for ribosomal proteins of RNA polymerase subunits), components of many functional systems appear to be fully compatible. One could consider this a moot point given the successful expression of eukaryotic proteins in bacteria routinely exploited in the laboratory. This parallel is not really valid, however, because in order to be fixed in the bacterial population and retained in the long term, a eukaryotic gene must confer selective advantage on the recipient bacterium. This is particularly pertinent for xenologous gene displacement because in these cases, the transferred, heterologous version of a gene should immediately become superior, from the standpoint of selection, to the original version typical of the recipient species. In one case, that of eukaryotic isoleucyl-tRNA synthetase displacing the original gene in some bacteria, this has been convincingly explained by acquisition of antibiotic resistance. It seems likely that these observations have general implications for xenologous gene displacement. In some cases of acquisition of new genes, the nature of the selective advantage also appears clear, such as for the ATP/ADP translocases acquired by intracellular parasitic bacteria, Chlamydia and Rickettsia. In most instances, however, comparative genomics can only point to the genes that have probably entered the given genome by horizontal transfer. Understanding the biological significance of horizontal gene transfer will require direct experimental studies with these genes.
Acknowledgments
We wish to offer our apologies and express our appreciation to all researchers whose important work, particularly on individual cases of probable horizontal gene transfer, is not cited here because of inevitable space restrictions. We thank A.S. Kondrashov for useful discussions, M.Y. Galperin and V. Anantharaman for sharing unpublished observations, and Y.I. Wolf for help with some of the computational analyses. Kira Makarova is supported by grants DE-FG02-98ER62583, DE-FG02-97ER62492, and DE-FG07-97ER20293 from the U.S. Department of Energy.
Visit the Annual Reviews home page at www.AnnualReviews.org
Literature Cited
- 1.
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. [PMC free article: PMC146917] [PubMed: 9254694]
- 2.
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–40. [PubMed: 9823893]
- 3.
- Aravind L, Koonin EV. The fukutin protein family–predicted enzymes modifying cell-surface molecules. Curr. Biol. 1999;9:R836–37. [PubMed: 10574772]
- 4.
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–44. [PubMed: 9825671]
- 5.
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Reply. Archaeal and bacterial hyperthermophiles: horizontal gene exchange or common ancestry? Trends Genet. 1999;15:299–300. [PubMed: 10431189]
- 6.
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–42. [PMC free article: PMC148307] [PubMed: 9973609]
- 7.
- Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944;149:297–326. [PMC free article: PMC2184805] [PubMed: 33226]
- 8.
- Barry CED, Brickman TJ, Hackstadt T. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol. Microbiol. 1993;9:273–83. [PubMed: 8412680]
- 9.
- Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. [PMC free article: PMC309208] [PubMed: 8451181]
- 10.
- Brochier C, Philippe H, Moreira D. The evolutionary history of ribosomal protein RpS14: horizontal gene transfer at the heart of the ribosome. Trends Genet. 2000;16:529–33. [PubMed: 11102698]
- 11.
- Brown JK. Bootstrap hypothesis tests for evolutionary trees and other dendrograms. Proc. Natl. Acad. Sci. USA. 1994;91:12293–97. [PMC free article: PMC45423] [PubMed: 7991621]
- 12.
- Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 1997;61:456–502. [PMC free article: PMC232621] [PubMed: 9409149]
- 13.
- Brown JR, Zhang J, Hodgson JE. A bacterial antibiotic resistance gene with eukaryotic origins. Curr. Biol. 1998;8:R365–67. [PubMed: 9635179]
- 14.
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L. et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–60. [PubMed: 8980231]
- 15.
- Cavalli-Sforza LL. The DNA revolution in population genetics. Trends Genet. 1998;14:60–65. [PubMed: 9520599]
- 16.
- Curnow AW, Hong K, Yuan R, Kim S, Martins O. et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA. 1997;94:11819–26. [PMC free article: PMC23611] [PubMed: 9342321]
- 17.
- Curnow AW, Tumbula DL, Pelaschier JT, Min B, Soll D. Glutamyl-tRNA (Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA. 1998;95:12838–43. [PMC free article: PMC23620] [PubMed: 9789001]
- 18.
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–28. [PubMed: 9787636]
- 19.
- Davie JR, Spencer VA. Signal transduction pathways and the modification of chromatin structure. Prog. Nucleic Acid Res. Mol. Biol. 2000;65:299–340. [PubMed: 11008491]
- 20.
- Doolittle RF, Handy J. Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr. Opin. Genet. Dev. 1998;8:630–36. [PubMed: 9914200]
- 21.
- Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–8. [PubMed: 10611671]
- 22.
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–29. [PubMed: 10381871]
- 23.
- Doolittle WF. Uprooting the tree of life. Sci. Am. 2000;282:90–95. [PubMed: 10710791]
- 24.
- Doolittle WF, Logsdon JM Jr. Archaeal genomics: do archaea have a mixed heritage? Curr. Biol. 1998;8:R209–11. [PubMed: 9512414]
- 25.
- Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 1997;51:527–64. [PubMed: 9343359]
- 26.
- Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA. 1996;93:13429–34. [PMC free article: PMC24110] [PubMed: 8917608]
- 27.
- Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. [PMC free article: PMC3158673] [PubMed: 10606811]
- 28.
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–27. [PubMed: 8743697]
- 29.
- Gaasterland T, Ragan MA. Microbial genescapes: phyletic and functional patterns of ORF distribution among prokaryotes. Microb. Comp. Genomics. 1998;3:199–217. [PubMed: 10027190]
- 30.
- Galperin MY, Koonin EV. Who’s your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 2000;18:609–13. [PubMed: 10835597]
- 31.
- Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10:1719–25. [PMC free article: PMC310969] [PubMed: 11076857]
- 32.
- Gogarten JP. Which is the most conserved group of proteins? Homology-orthology, paralogy, xenology, and the fusion of independent lineages. J. Mol. Evol. 1994;39:541–43. [PubMed: 7807544]
- 33.
- Golding GB, Gupta RS. Protein-based phylogenies support a chimeric origin for the eukaryotic genome. Mol. Biol. Evol. 1995;12:1–6. [PubMed: 7877484]
- 34.
- Gould SJ. The exaptive excellence of spandrels as a term and prototype. Proc. Natl. Acad. Sci. USA. 1997;94:10750–55. [PMC free article: PMC23474] [PubMed: 11038582]
- 35.
- Grantham R, Gautier C, Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980;8:1893–912. [PMC free article: PMC324046] [PubMed: 6159596]
- 36.
- Grantham R, Gautier C, Gouy M, Mercier R, Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:R49–62. [PMC free article: PMC327256] [PubMed: 6986610]
- 37.
- Gray MW. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 1992;141:233–357. [PubMed: 1452433]
- 38.
- Gray MW. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 1999;9:678–87. [PubMed: 10607615]
- 39.
- Groisman EA, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–49. [PubMed: 9294889]
- 40.
- Gupta RS, Golding GB. The origin of the eukaryotic cell. Trends Biochem. Sci. 1996;21:166–71. [PubMed: 8871398]
- 41.
- Handy J, Doolittle RF. An attempt to pinpoint the phylogenetic introduction of glutaminyl-tRNA synthetase among bacteria. J. Mol. Evol. 1999;49:709–15. [PubMed: 10594171]
- 42.
- Hartl DL, Dykhuizen DE, Berg DE. Accessory DNAs in the bacterial gene pool: playground for coevolution. Ciba Found. Symp. 1984;102:233–45. [PubMed: 6319094]
- 43.
- Hilario E, Gogarten JP. Horizontal transfer of ATPase genes—the tree of life becomes a net of life. Biosystems. 1993;31:111–19. [PubMed: 8155843]
- 44.
- Hopwood DA, Chater KF, Bibb MJ. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology. 1995;28:65–102. [PubMed: 8688641]
- 45.
- Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–27. [PubMed: 10647936]
- 46.
- Ibba M, Curnow AW, Soll D. Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem. Sci. 1997;22:39–42. [PubMed: 9048478]
- 47.
- Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 1999;16:332–46. [PubMed: 10331260]
- 48.
- Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA. 1999;96:3801–6. [PMC free article: PMC22375] [PubMed: 10097118]
- 49.
- Jeanthon C. Molecular ecology of hydrothermal vent microbial communities. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2000;77:117–33. [PubMed: 10768471]
- 50.
- Kaneko T, Tabata S. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 1997;38:1171–76. [PubMed: 9435137]
- 51.
- Knopf CW. Evolution of viral DNA-dependent DNA polymerases. Virus Genes. 1998;16:47–58. [PubMed: 9562890]
- 52.
- Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 1999;9:649–56. [PubMed: 10607611]
- 53.
- Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–76. [PubMed: 10892642]
- 54.
- Koonin EV, Mushegian AR. Complete genome sequences of cellular life forms: glimpses of theoretical evolutionary genomics. Curr. Opin. Genet. Dev. 1996;6:757–62. [PubMed: 8994848]
- 55.
- Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–36. [PubMed: 8855656]
- 56.
- Koonin EV, Mushegian AR, Galperin MY, Walker DR. Comparison of archaeal and bacterial genomes: Computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol. 1997;25:619–37. [PubMed: 9379893]
- 57.
- Kyrpides NC, Olsen GJ. 1999. Archaeal and bacterial hyperthermophiles: horizontal gene exchange or common ancestry?Trends Genet. 15:298–99. [PubMed: 10431189]
- 58.
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999;33:351–97. [PubMed: 10690412]
- 59.
- Lawrence JG. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–59. [PubMed: 9294891]
- 60.
- Lawrence JG. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 1999;9:642–48. [PubMed: 10607610]
- 61.
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–97. [PubMed: 9089078]
- 62.
- Lengeler KB, Davidson RC, D’Souza C, Harashima T, Shen WC. et al. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 2000;64:746–85. [PMC free article: PMC99013] [PubMed: 11104818]
- 63.
- Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8:1038–47. [PubMed: 9799791]
- 64.
- Li J, Lee GI, Van Doren SR, Walker JC. The FHA domain mediates phosphoprotein interactions. J. Cell Sci. 2000;113:4143–49. [PubMed: 11069759]
- 65.
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. [PubMed: 10094048]
- 66.
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–77. [PMC free article: PMC85410] [PubMed: 10713161]
- 67.
- Logsdon JM, Faguy DM. Thermotoga heats up lateral gene transfer. Curr. Biol. 1999;9:R747–51. [PubMed: 10531001]
- 68.
- Longhi S, Cambillau C. Structure-activity of cutinase, a small lipolytic enzyme. Biochim. Biophys. Acta. 1999;1441:185–96. [PubMed: 10570246]
- 69.
- Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994;58:563–602. [PMC free article: PMC372978] [PubMed: 7968924]
- 70.
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL. et al. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–28. [PubMed: 10413400]
- 71.
- Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 2000;25:50–52. [PubMed: 10664582]
- 72.
- Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW. et al. The genome of the extremely radioresis-tant bacterium Deinococcus radiodurans: comparative genomics. Microbiol. Mol. Biol. Rev. 2001;65:44–79. [PMC free article: PMC99018] [PubMed: 11238985]
- 73.
- Margulis L, Chase D, To LP. Possible evolutionary significance of spirochaetes. Proc. R. Soc. London Ser. B. 1979;204:189–98. [PubMed: 36621]
- 74.
- Martin W. Mosaic bacterial chromosomes: a challenge en route to a tree of genomes. BioEssays. 1999;21:99–104. [PubMed: 10193183]
- 75.
- Martin W, Herrmann RG. 1998. Gene transfer from organelles to the nucleus: how much, what happens, and why?Plant Physiol. 118:9–17. [PMC free article: PMC1539188] [PubMed: 9733521]
- 76.
- Medigue C, Rouxel T, Vigier P, Henaut A, Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J. Mol. Biol. 1991;222:851–56. [PubMed: 1762151]
- 77.
- Moreira D. Multiple independent horizontal transfers of informational genes from bacteria to plasmids and phages: implications for the origin of bacterial replication machinery. Mol. Microbiol. 2000;35:1–5. [PubMed: 10632872]
- 78.
- Moreira D, Philippe H. Molecular phylogeny: pitfalls and progress. Int. Microbiol. 2000;3:9–16. [PubMed: 10963328]
- 79.
- Moyer CL, Dobbs FC, Karl DM. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 1995;61:1555–62. [PMC free article: PMC167411] [PubMed: 7538279]
- 80.
- Mrazek J, Karlin S. Detecting alien genes in bacterial genomes. Ann. NY Acad. Sci. 1999;870:314–29. [PubMed: 10415493]
- 81.
- Mushegian AR, Koonin EV. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–90. [PubMed: 8783936]
- 82.
- Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–99. [PubMed: 7846533]
- 83.
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ. et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–29. [PubMed: 10360571]
- 84.
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. [PubMed: 10830951]
- 85.
- Olendzenski L, Liu L, Zhaxybayeva O, Murphey R, Shin DG, Gogarten JP. Horizontal transfer of archaeal genes into the Deinococcaceae: detection by molecular and computer-based approaches. J. Mol. Evol. 2000;51:587–99. [PubMed: 11116332]
- 86.
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. [PubMed: 9115194]
- 87.
- Paerl HW. Microscale physiological and ecological studies of aquatic cyanobacteria: macroscale implications. Microsc. Res. Technol. 1996;33:47–72. [PubMed: 8820664]
- 88.
- Patterson C. Homology in classical and molecular biology. Mol. Biol. Evol. 1988;5:603–25. [PubMed: 3065587]
- 89.
- Pennisi E. Genome data shake tree of life. Science. 1998;280:672–74. [PubMed: 9599142]
- 90.
- Pennisi E. 1999. Is it time to uproot the tree of life?Science 284:1305–7. [PubMed: 10383313]
- 91.
- Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 1999;289:729–45. [PubMed: 10369758]
- 92.
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–99. [PubMed: 10949293]
- 93.
- Risatti JB, Capman WC, Stahl DA. Community structure of a microbial mat: the phylogenetic dimension. Proc. Natl. Acad. Sci. USA. 1994;91:10173–77. [PMC free article: PMC44980] [PubMed: 7937858]
- 94.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–25. [PubMed: 3447015]
- 95.
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–34. [PMC free article: PMC102444] [PubMed: 10592234]
- 96.
- Sicot FX, Mesnage M, Masselot M, Exposito JY, Garrone R. et al. Molecular adaptation to an extreme environment: origin of the thermal stability of the pompeii worm collagen. J. Mol. Biol. 2000;302:811–20. [PubMed: 10993725]
- 97.
- Smith MW, Feng DF, Doolittle RF. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem. Sci. 1992;17:489–93. [PubMed: 1471257]
- 98.
- Sprague GF Jr. Genetic exchange between kingdoms. Curr. Opin. Genet. Dev. 1991;1:530–33. [PubMed: 1822285]
- 99.
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R. et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–59. [PubMed: 9784136]
- 100.
- Subramanian G, Koonin EV, Aravind L. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum. Infect. Immun. 2000;68:1633–48. [PMC free article: PMC97324] [PubMed: 10678983]
- 101.
- Sundstrom L. The potential of integrons and connected programmed rearrangements for mediating horizontal gene transfer. APMIS Suppl. 1998;84:37–42. [PubMed: 9850680]
- 102.
- Syvanen M. Cross-species gene transfer; implications for a new theory of evolution. J. Theor. Biol. 1985;112:333–43. [PubMed: 2984477]
- 103.
- Syvanen M. Horizontal gene transfer: evidence and possible consequences. Annu. Rev. Genet. 1994;28:237–61. [PubMed: 7893125]
- 104.
- Tannock GW. The intestinal microflora: potentially fertile ground for microbial physiologists. Adv. Microb. Physiol. 2000;42:25–46. [PubMed: 10907550]
- 105.
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–37. [PubMed: 9381173]
- 106.
- Tatusov RL, Mushegian AR, Bork P, Brown NP, Hayes WS. et al. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr. Biol. 1996;6:279–91. [PubMed: 8805245]
- 107.
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT. et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. [PMC free article: PMC29819] [PubMed: 11125040]
- 108.
- Tatusova TA, Karsch-Mizrachi I, Ostell JA. Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics. 1999;15:536–43. [PubMed: 10487861]
- 109.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. [PMC free article: PMC308517] [PubMed: 7984417]
- 110.
- Walker DR, Koonin EV. SEALS: a system for easy analysis of lots of sequences. Intell. Syst. Mol. Biol. 1997;5:333–39. [PubMed: 9322058]
- 111.
- Ward DM, Ferris MJ, Nold SC, Bateson MM. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 1998;62:1353–70. [PMC free article: PMC98949] [PubMed: 9841675]
- 112.
- White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD. et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–77. [PMC free article: PMC4147723] [PubMed: 10567266]
- 113.
- Winkler HH, Neuhaus HE. Non-mitochondrial ATP transport. Trends Biochem. Sci. 1999;24:64–68. [PubMed: 10098400]
- 114.
- Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–36. [PMC free article: PMC98992] [PubMed: 10704480]
- 115.
- Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed: 10447505]
- 116.
- Wolf YI, Aravind L, Koonin EV. Rickettsiae and Chlamydiae: evidence of horizontal gene transfer and gene exchange. Trends Genet. 1999;15:173–75. [PubMed: 10322483]
- 117.
- Wolf YI, Kondrashov AS, Koonin EV. 2000. Interkingdom gene fusions. Genome Biol. 1:research0013.1–0013.15. [PMC free article: PMC16144] [PubMed: 11178267]
- 118.
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 2001;11:356–72. [PubMed: 11230160]
- 119.
- Worning P, Jensen LJ, Nelson KE, Brunak S, Ussery DW. Structural analysis of DNA sequence: evidence for lateral gene transfer in Thermotoga maritima. Nucleic Acids Res. 2000;28:706–9. [PMC free article: PMC102551] [PubMed: 10637321]
Publication Details
Author Information and Affiliations
Authors
Eugene V. Koonin,1 Kira S. Makarova,1,2 and L. Aravind2.Affiliations
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Koonin EV, Makarova KS, Aravind L. Horizontal Gene Transfer in Prokaryotes: Quantification and Classification. In: Annual Reviews Collection [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002 Nov.