By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002.

Molecular Biology of the Cell. 4th edition.
Show details- α helix
see alpha helix
- ABC transporter proteins
Large superfamily of membrane transport proteins that use the energy of hydrolysis of ATP to transfer peptides and a variety of small molecules across membranes.
- acetyl
Chemical group derived from acetic acid. Acetyl groups are important in metabolism and are added covalently to some proteins as a posttranslational modification.
- acetyl CoA
Small water-soluble molecule that carries acetyl groups in cells. It consists of an acetyl group linked to coenzyme A (CoA) by an easily hydrolyzable thioester bond. (See Figure 2–62.)
- acetylcholine receptor
Ion channel that opens in response to binding of acetylcholine, thereby converting a chemical signal into an electrical one. Best understood example of a transmitter-gated channel. Sometimes called the nicotinic acetylcholine receptor to distinguish it from a muscarinic acetylcholine receptor, which is a G-protein-linked cell-surface receptor.
- acetylcholine
Neurotransmitter that functions at a class of chemical synapses known as cholinergic synapses. Found both in the brain and in the peripheral nervous system. It is the neurotransmitter at vertebrate neuromuscular junctions. (See Figure 15–9.)
- acid
Substance that releases protons when dissolved in water, forming a hydronium ion (H3O+).
- acid hydrolase
Any of a group of diverse hydrolytic enzymes (including proteases, nucleases, glycosidases, etc.) that have their optimal activity at acid pH (around 5.0) and are found in lysosomes.
- acquired immunological tolerance
Unresponsiveness of the immune system to a given foreign antigen that can develop in some circumstances.
- acrosomal vesicle
Region at the head end of a sperm cell that contains a sac of hydrolytic enzymes used to digest the protective coating of the egg.
- acrosome reaction
Reaction that occurs when a sperm starts to enter an egg, in which the contents of the acrosomal vesicle are released, helping the sperm to penetrate the zona pellucida.
- actin
Abundant protein that forms actin filaments in all eucaryotic cells. The monomeric form is sometimes called globular or G-actin; the polymeric form is filamentous or F-actin.
- actin-binding protein
Protein that associates with either actin monomers or actin filaments in cells and modifies their properties. Examples include myosin, α-actinin, and profilin.
- actin filament (microfilament)
Helical protein filament formed by the polymerization of globular actin molecules. A major constituent of the cytoskeleton of all eucaryotic cells and part of the contractile apparatus of skeletal muscle. (See Panel 16–1, p. 909.)
- action potential
Rapid, transient, self-propagating electrical excitation in the plasma membrane of a cell such as a neuron or muscle cell. Action potentials, or nerve impulses, make possible long-distance signaling in the nervous system.
- activated carrier
Small diffusible molecule in cells that stores easily-exchangeable energy in the form of one or more energy-rich covalent bonds. Examples are ATP and NADPH. Also called a coenzyme.
- activation energy
Extra energy that must be possessed by atoms or molecules in addition to their ground-state energy in order to undergo a particular chemical reaction. (See Figure 9–1.)
- active site
Region of an enzyme surface to which a substrate molecule binds in order to undergo a catalyzed reaction.
- active transport
Movement of a molecule across a membrane or other barrier driven by energy other than that stored in the electrochemical gradient of the transported molecule.
- acyl group
Functional group derived from a carboxylic acid (
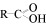 ). (R represents an alkyl group, such as methyl.)
). (R represents an alkyl group, such as methyl.)- adaptation
Adjustment of sensitivity following repeated stimulation. This is the mechanism that allows a neuron, a photodetector, or a bacterium to react to small changes in stimuli even against a high background level of stimulation.
- adaptin
Protein that binds clathrin to the membrane surface in clathrin-coated vesicles.
- adaptive immune response
Response of the vertebrate immune system to a specific antigen that typically generates immunological memory.
- adaptor protein
General term for proteins in intracellular signaling pathways that link different proteins in the pathway directly together.
- adenomatous polyposis coli (APC)
Tumor suppressor protein that forms part of a protein complex that recruits free cytoplasmic β-catenin and degrades it.
- adenosine triphosphate
see ATP
- adenylyl cyclase (adenylate cyclase)
Membrane-bound enzyme that catalyzes the formation of cyclic AMP from ATP. An important component of some intracellular signaling pathways.
- adherens junction
Cell junction in which the cytoplasmic face of the plasma membrane is attached to actin filaments. Examples include the adhesion belts linking adjacent epithelial cells and the focal contacts on the lower surface of cultured fibroblasts.
- adhesion belt
Beltlike adherens junction that encircles the apical end of an epithelial cell and attaches it to the adjoining cell. Also known as the zonula adherens.
- adhesion plaque
see focal adhesion.
- adipocyte
A fat cell.
- ADP (adenosine 5-diphosphate)
Nucleotide that is produced by hydrolysis of the terminal phosphate of ATP. It regenerates ATP when phosphorylated by an energy-generating process such as oxidative phosphorylation. (See Figure 2–57.)
- adrenaline (epinephrine)
Hormone released by chromaffin cells (in the adrenal gland) and by some neurons in response to stress. Produces “fight or flight” responses, including increased heart rate and blood sugar levels.
- aerobic
Describes a process that requires, or occurs in the presence of, gaseous oxygen (O2).
- affinity chromatography
Type of chromatography in which the protein mixture to be purified is passed over a matrix to which specific ligands for the required protein are attached, so that the protein is retained on the matrix.
- affinity constant (association constant) (Ka)
Measure of the strength of binding of the components in a complex. For components A and B and a binding equilibrium A + B ⇌ AB, the association constant is given by [AB]/[A][B], and is larger the tighter the binding between A and B. (See also dissociation constant.)
- affinity maturation
Progressive increase in the affinity of antibodies for the immunizing antigen with the passage of time after immunization.
- Akt
see protein kinase B
- alcohol
Polar organic molecule that contains a functional hydroxyl group (–OH) bound to a carbon atom that is not in an aromatic ring. An example is ethyl alcohol (CH3CH2OH).
- aldehyde
Organic compound that contains the group. An example is glyceraldehyde. Can be oxidized to an acid or reduced to an alcohol.
- alga (algae)
Informal term used to describe a wide range of simple unicellular and multicellular eucaryotic photosynthetic organisms. Examples include Nitella, Volvox, and Fucus.
- alkaloid
Small but chemically complex nitrogen-containing metabolite produced by plants as a defense against herbivores. Examples include caffeine, morphine, and colchicine.
- alkane (adjective aliphatic)
Compound of carbon and hydrogen that has only single covalent bonds. An example is ethane (CH3CH3).
- alkene
Hydrocarbon with one or more carbon-carbon double bonds. An example is ethylene (CH2CH2).
- alkyl group
General term for a group of covalently linked carbon and hydrogen atoms such as methyl (–CH3) or ethyl (–CH2CH3) groups. These groups can be formed by removing a hydrogen atom from an alkane.
- allele
One of a set of alternative forms of a gene. In a diploid cell each gene will have two alleles, each occupying the same position (locus) on homologous chromosomes.
- allelic exclusion
The expression of an immunoglobulin chain (or T cell receptor chain) gene from only one of the two homologous loci present for that gene in the lymphocyte.
- allosteric protein
Protein that changes from one conformation to another when it binds another molecule or when it is covalently modified. The change in conformation alters the activity of the protein and can form the basis of directed movement.
- alpha helix ( helix)
Common folding pattern in proteins in which a linear sequence of amino acids folds into a right-handed helix stabilized by internal hydrogen bonding between backbone atoms.
- alternative RNA splicing
The production of different proteins from the same RNA transcript by splicing it in different ways.
- amide
Molecule containing a carbonyl group linked to an amine.
- amine
Chemical group containing nitrogen and hydrogen. It becomes positively charged in water.
- amino acid
Organic molecule containing both an amino group and a carboxyl group. Those that serve as the building blocks of proteins are alpha amino acids, having both the amino and carboxyl groups linked to the same carbon atom. (See Panel 3–1, pp. 132–133.)
- aminoacyl-tRNA synthetase
Enzyme that attaches the correct amino acid to a tRNA molecule to form an aminoacyl-tRNA. (See Figure 6–57.)
- amino group
Weakly basic functional group derived from ammonia (NH3) in which one or more hydrogen atoms are replaced by another atom. In aqueous solution it can accept a proton and carry a positive charge.
- amino terminus (N terminus)
The end of a polypeptide chain that carries a free α-amino group.
- aminoacyl tRNA
Activated form of amino acid used in protein synthesis. Consists of an amino acid linked through a labile ester bond from its carboxyl group to a hydroxyl group on tRNA. (See Figure 6–57.)
- AMP (adenosine 5-monophosphate)
One of the four nucleotides in an RNA molecule. Two phosphates are added to AMP to form ATP. (See Panel 2–6, pp. 120–121.)
- amphipathic
Having both hydrophobic and hydrophilic regions, as in a phospholipid or a detergent molecule.
- anabolism
System of biosynthetic reactions in a cell by which large molecules are made from smaller ones.
- anaerobic
Describes a cell, organism, or metabolic process that functions in the absence of air or, more precisely, in the absence of molecular oxygen (O2).
- anaphase
Stage of mitosis during which the two sets of chromosomes separate and move away from each other. Composed of anaphase A (chromosomes move toward the two spindle poles) and anaphase B (spindle poles move apart).
- anaphase-promoting complex (APC)
Ubiquitin ligase that promotes the destruction of a set of proteins, some of which initiate the separation of sister chromatids during the metaphase-to-anaphase transition during mitosis.
- anchorage dependence
Dependence of cell growth on attachment to a substratum.
- anchoring junction
Type of cell junction that attaches cells to neighboring cells or to the extracellular matrix.
- angiogenesis
Growth of new blood vessels by sprouting from existing ones.
- Ångstrom ()
Unit of length used to measure atoms and molecules. Equal to 10–10 meter or 0.1 nanometer (nm).
- animal pole
In yolky eggs, that end free of yolk that cleaves more rapidly than the vegetal pole.
- ankyrin
Protein mainly responsible for attaching the spectrin cytoskeleton to the red blood cell plasma membrane.
- antenna complex
Part of a photosystem that captures light energy and channels it into the photochemical reaction center. It consists of protein complexes that bind large numbers of chlorophyll molecules and other pigments.
- anterior
Situated toward the head end of the body.
- anteroposterior
Describes the axis running from the head to the tail of the animal body.
- antibiotic
Substance such as penicillin or streptomycin that is toxic to microorganisms. Usually a product of a particular microorganism or plant.
- antibody (immunoglobulin)
Protein produced by B cells in response to a foreign molecule or invading microorganism. Often binds to the foreign molecule or cell extremely tightly, thereby inactivating it or marking it for destruction by phagocytosis or complement-induced lysis.
- anticodon
Sequence of three nucleotides in a transfer RNA molecule that is complementary to a three-nucleotide codon in a messenger RNA molecule.
- antigen
Molecule that is able to provoke an immune response.
- antigenic determinant (epitope)
Specific region of an antigenic molecule that binds to an antibody or a T cell receptor.
- antigenic variation
The ability to change the antigens displayed on the cell surface; a property of some pathogenic microorganisms that enables them to evade attack by the immune system.
- antigen-presenting cell
Cell that displays foreign antigen complexed with MHC molecules on its surface.
- antiparallel
Describes the relative orientation of the two strands in a DNA double helix; the polarity of one strand is oriented in the opposite direction to that of the other.
- antiporter
Carrier protein that transports two different ions or small molecules across a membrane in opposite directions, either simultaneously or in sequence.
- antisense RNA
RNA complementary to a specific RNA transcript of a gene that can hybridize to the specific RNA and block its function.
- APC
- apical
Describes the tip of a cell, a structure, or an organ. The apical surface of an epithelial cell is the exposed free surface, opposite to the basal surface. The basal surface rests on the basal lamina that separates the epithelium from other tissue.
- apoptosis
Form of cell death, also known as programmed cell death, in which a ‘suicide’ program is activated within the cell, leading to fragmentation of the DNA, shrinkage of the cytoplasm, membrane changes and cell death without lysis or damage to neighboring cells. It is a normal phenomenon, occurring frequently in a multicellular organism.
- aqueous
Pertaining to water, as for example, in an aqueous solution.
- archea (singular archeon)
Members of one of the two major divisions of procaryotes (the Archea), the other being the Bacteria.
- ARF protein
Monomeric GTPase responsible for regulating both COPI coat assembly and clathrin coat assembly at Golgi membranes.
- aromatic
Describes a molecule that contains carbon atoms in a ring, commonly drawn as linked through alternating single and double bonds. Often a molecule related to benzene.
- ARP complex (ARP2/3 complex)
Complex of proteins that nucleates actin filament growth from the minus end.
- asexual reproduction
Any type of reproduction (such as budding in Hydra, binary fission in bacteria, or mitotic division in eucaryotic microorganisms) that does not involve gamete formation and fusion. It produces an individual genetically identical to the parent.
- association constant
see affinity constant
- aster
Star-shaped system of microtubules emanating from a centrosome or from a pole of a mitotic spindle.
- astral microtubule
In the mitotic spindle, any of the microtubules radiating from the aster which are not attached to a kinetochore of a chromosome.
- asymmetric cell division
Cell division that produces two daughter cells that differ, for example in size or in the presence or absence of some cytoplasmic constituent.
- atomic weight
Mass of an atom relative to the mass of a hydrogen atom. Essentially equal to the number of protons plus neutrons.
- ATP (adenosine 5-triphosphate)
Nucleoside triphosphate composed of adenine, ribose, and three phosphate groups that is the principal carrier of chemical energy in cells. The terminal phosphate groups are highly reactive in the sense that their hydrolysis, or transfer to another molecule, takes place with release of a large amount of free energy. (See Figure 2–26.)
- ATP synthase
Enzyme complex in the inner membrane of a mitochon-drion and the thylakoid membrane of a chloroplast that catalyzes the formation of ATP from ADP and inorganic phosphate during oxidative phosphorylation and photosynthesis, respectively. Also present in the plasma membrane of bacteria.
- ATPase
Enzyme that catalyzes a process involving the hydrolysis of ATP. A large number of different proteins have ATPase activity.
- autoantibody
Antibody produced by an individual against a protein, or other potential antigen, of its own cells and tissues. Autoantibodies can cause autoimmune disease.
- autocatalysis
Reaction that is catalyzed by one of its products, creating a positive feedback (self-amplifying) effect on the reaction rate.
- autocrine signaling
Type of cell signaling in which a cell secretes signal molecules that act on itself or on other adjacent cells of the same type.
- autoimmune disease
A pathological state in which the body mounts an immune response against one or more of its own potential antigens.
- autophagy
Digestion of worn-out organelles by the cell’s own lysosomes.
- autoradiography
Technique in which a radioactive object produces an image of itself on a photographic film. The image is called an autoradiograph or autoradiogram.
- autosome
Any chromosome other than a sex chromosome.
- avidity
Total binding strength of a polyvalent antibody with a polyvalent antigen.
- Avogadro’s number
6 × 1023. This is the number of atoms in 1 gram of hydrogen, and thus in the atomic or molecular weight equivalent in grams of any element or molecule.
- axon
Long nerve cell process that is capable of rapidly conducting nerve impulses over long distances so as to deliver signals to other cells.
- axonal transport
Directed transport of organelles and molecules along a nerve cell axon. It can be anterograde (outward from the cell body) or retrograde (back toward the cell body).
- axoneme
Bundle of microtubules and associated proteins that forms the core of a cilium or flagellum in a eucaryotic cell and is responsible for their movements.
- β sheet
see beta sheet
- B cell (B lymphocyte)
Type of lymphocyte that makes antibodies.
- bacteria (singular bacterium)
Members of the Bacteria, one of the two major divisions of procaryotes, the other being the Archea. Most exist as single cells and some cause disease.
- bacterial artificial chromosome (BAC)
Cloning vector that can accommodate large pieces of DNA up to 1 million base pairs.
- bacteriophage (phage)
Any virus that infects bacteria. Bacteriophages were the first entities used for the study of molecular genetics and are now widely used as cloning vectors.
- bacteriorhodopsin
Pigmented protein found in the plasma membrane of a salt-loving bacterium, Halobacterium halobium. It pumps protons out of the cell in response to light.
- basal
Situated near the base. The basal surface of a cell is opposite the apical surface.
- basal body
Short cylindrical array of microtubules plus their associated proteins found at the base of a eucaryotic cell cilium or flagellum. Serves as a nucleation site for the growth of the axoneme. Closely similar in structure to a centriole.
- basal lamina (basal laminae)
Thin mat of extracellular matrix that separates epithelial sheets, and many other types of cells such as muscle or fat cells, from connective tissue.
- base
A substance that can accept a proton in solution. The purines and pyrimidines in DNA and RNA are organic nitrogenous bases and are often referred to simply as bases.
- base pair
Two nucleotides in an RNA or DNA molecule that are held together by hydrogen bonds—for example, G pairs with C, and A with T or U.
- basic
Having the properties of a base.
- benign
Describes tumors that are self-limiting in their growth and noninvasive.
- benzene
Molecule composed of a six-membered ring of carbon atoms, commonly drawn containing three alternating double bonds. The benzene ring occurs as part of many biological molecules.
- beta-catenin (-catenin)
Multifunctional cytoplasmic protein that is involved in cadherin-mediated cell–cell adhesion, linking cadherins to the actin cytoskeleton. Can also act independently as a gene regulatory protein. Has an important role in animal development as part of a Wnt signaling pathway.
- beta sheet ( sheet)
Common structural motif in proteins in which different sections of the polypeptide chain run alongside each other, joined together by hydrogen bonding between atoms of the polypeptide backbone. Also known as a β-pleated sheet.
- binding site
A region on the surface of one molecule (usually a protein or nucleic acid) that can interact with another molecule through noncovalent bonding.
- biosphere
The world of living organisms.
- biotin
Low-molecular-weight compound used as a coenzyme. Useful technically as a covalent label for proteins, allowing them to be detected by the egg protein avidin, which binds extremely tightly to biotin. (See Figure 2–63.)
- bivalent
A duplicated chromosome paired with its homologous duplicated chromosome at the beginning of meiosis.
- black membrane
Artificial planar lipid bilayer membrane.
- blastomere
One of the cells formed by the cleavage of a fertilized egg.
- blastula
Early stage of an animal embryo, usually consisting of a hollow ball of cells, before gastrulation begins.
- blotting
Biochemical technique in which macromolecules separated on an agarose or polyacrylamide gel are transferred to a nylon membrane or sheet of paper, thereby immobilizing them for further analysis. (See Northern blotting, Southern blotting, Western blotting.)
- bond energy
Strength of the chemical linkage between two atoms, measured by the energy in kilocalories or kilojoules needed to break it.
- bright-field microscope
The normal light microscope in which the image is obtained by simple transmission of light through the object being viewed.
- brush border
Dense covering of microvilli on the apical surface of epithelial cells in the intestine and kidney. The microvilli aid absorption by increasing the surface area of the cell.
- budding yeast
Common name often given to the baker’s yeast Saccharomyces cerevisiae, a common experimental organism, which divides by budding off a smaller cell.
- C terminus
see carboxyl terminus
- Ca2+/calmodulin-dependent protein kinase (CaM kinase)
Protein kinase whose activity is regulated by the binding of Ca2+-activated calmodulin (Ca2+/calmodulin), and which indirectly mediates the effects of Ca2+ by phosphorylation of other proteins.
- cadherin
A member of a family of proteins that mediates Ca2+-dependent cell–cell adhesion in animal tissues.
- caged molecule
Organic molecule designed to change into an active form when irradiated with light of a specific wavelength. An example is caged ATP.
- calcium pump (Ca2+ ATPase)
Transport protein in the membrane of the sarcoplasmic reticulum of muscle cells (and elsewhere) that pumps Ca2+ out of the cytoplasm into the sarcoplasmic reticulum using the energy of ATP hydrolysis.
- calmodulin
Ubiquitous calcium-binding protein whose binding to other proteins is governed by changes in intracellular Ca2+ concentration. Its binding modifies the activity of many target enzymes and membrane transport proteins.
- calorie
Unit of heat. One calorie (small “c”) is the amount of heat needed to raise the temperature of 1 gram of water by 1°C. A kilocalorie (1000 calories) is the unit used to describe the energy content of foods.
- Calvin cycle (Calvin-Benson cycle)
Major metabolic pathway by which CO2 is incorporated into carbohydrate during the second stage of photosynthesis (carbon fixation) in plants. Also called the carbon-fixation cycle.
- CAM
- CaM kinase
see Ca2+/calmodulin-dependent protein kinase
- CaM-kinase II
Multifunctional Ca2+/calmodulin-dependent protein kinase found in all animal cells that undergoes autophosphorylation when activated. It is especially abundant in brain and is thought to have a role in learning and memory in vertebrates.
- cAMP
see cyclic AMP
- capacitation
Poorly understood process that sperm must go through in the female reproductive tract before they are competent for fertilization.
- capsid
Protein coat of a virus, formed by the self-assembly of one or more protein subunits into a geometrically regular structure.
- carbohydrate
General term for sugars and related compounds containing carbon, hydrogen, and oxygen, usually with the empirical formula (CH2O)n.
- carbon fixation
Process by which green plants incorporate carbon atoms from atmospheric carbon dioxide into sugars. The second stage of photosynthesis.
- carbon-fixation cycle
see Calvin cycle
- carbonyl group
Pair of atoms consisting of a carbon atom linked to an oxygen atom by a double bond (C=O).
- carboxyl group
Carbon atom linked both to an oxygen atom by a double bond and to a hydroxyl group. Molecules containing a carboxyl group are weak acids—carboxylic acids (
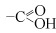 ).
).- carboxyl terminus (C terminus)
The end of a polypeptide chain that carries a free α-carbonyl group.
- carcinogen
Any agent, such as a chemical or a form of radiation, that causes cancer.
- carcinogenesis
Generation of a cancer.
- carcinoma
Cancer of epithelial cells. The most common form of human cancer.
- cardiac muscle
Specialized form of striated muscle found in the heart, consisting of individual heart muscle cells linked together by cell junctions.
- carrier protein
Membrane transport protein that binds to a solute and transports it across the membrane by undergoing a series of conformational changes.
- cartilage
Form of connective tissue composed of cells (chondrocytes) embedded in a matrix rich in type II collagen and chondroitin sulfate.
- caspase
Any of a family of intracellular proteases that are involved in initiating the cellular events of apoptosis.
- catabolism
General term for the enzyme-catalyzed reactions in a cell by which complex molecules are degraded to simpler ones with release of energy. Intermediates in these reactions are sometimes called catabolites.
- catalyst
Substance that can lower the activation energy of a reaction, thus increasing its rate.
- caveola (caveolae)
Invaginations at the cell surface that bud off internally to form pinocytic vesicles. Thought to form from lipid rafts, regions of membrane rich in certain lipids.
- CD28
Cell-surface protein on T cells that binds the co-stimulatory B7 protein on “professional” antigen-presenting cells, providing an additional signal required for the activation of a naïve T cell by antigen.
- CD4
Co-receptor protein found on helper T cells that binds to class II MHC molecules outside the antigen-binding site.
- CD8
Co-receptor protein found on cytotoxic T cells that binds to class I MHC molecules outside the antigen-binding site.
- cdc gene
see cell-division-cycle gene
- Cdk inhibitor protein (CKI)
Protein that binds to and inhibits cyclin-Cdk complexes, primarily involved in the control of G1 and S phases.
- Cdk-activating kinase (CAK)
Protein kinase that phosphorylates Cdks in cyclin-Cdk complexes, activating the Cdk.
- Cdk
see cyclin-dependent kinase
- cDNA
DNA molecule made as a copy of messenger RNA and therefore lacking the introns that are present in genomic DNA. cDNA clones represent DNA cloned from cDNA and a collection of such clones, usually representing the genes expressed in a particular cell type or tissue, is a cDNA library.
- cell adhesion molecule (CAM)
Protein on the surface of an animal cell that mediates cell–cell binding or cell–matrix binding.
- cell body
Main part of a nerve cell that contains the nucleus. The other parts are the axons and dendrites.
- cell coat
see glycocalyx
- cell cortex
Specialized layer of cytoplasm on the inner face of the plasma membrane. In animal cells it is an actin-rich layer responsible for movements of the cell surface.
- cell cycle (cell-division cycle)
Reproductive cycle of a cell: the orderly sequence of events by which a cell duplicates its contents and divides into two.
- cell division
Separation of a cell into two daughter cells. In eucaryotic cells it entails division of the nucleus (mitosis) closely followed by division of the cytoplasm (cytokinesis).
- cell fate
In developmental biology, describes what a particular cell at a given stage of development will normally give rise to.
- cell fusion
Process in which the plasma membranes of two cells fuse down at the point of contact between them, allowing the two cytoplasms to mingle.
- cell junction
Specialized region of connection between two cells or between a cell and the extracellular matrix.
- cell line
Population of cells of plant or animal origin capable of dividing indefinitely in culture.
- cell plate
Flattened membrane-bounded structure that forms by fusing vesicles in the cytoplasm of a dividing plant cell and is the precursor of the new cell wall.
- cell wall
Mechanically strong extracellular matrix deposited by a cell outside its plasma membrane. It is prominent in most plants, bacteria, algae, and fungi. Not present in most animal cells.
- cell-cycle control system
Network of regulatory proteins that governs progression of a eucaryotic cell through the cell cycle.
- cell-division-cycle gene (cdc gene)
Gene that controls a specific step or set of steps in the eucaryotic cell cycle. Originally identified in yeasts.
- cell-free system
Fractionated cell homogenate that retains a particular biological function of the intact cell, and in which biochemical reactions and cell processes can be more easily studied.
- cell-mediated immune response
That part of an adaptive immune response in which antigen-specific T cells are activated to perform various functions such as killing infected cells and activating macrophages.
- cellularization
The formation of cells around each nucleus in a multinucleate cytoplasm, transforming it into a multicellular structure.
- cellulose
Structural polysaccharide consisting of long chains of covalently linked glucose units. It provides tensile strength in plant cell walls.
- centimorgan
see genetic map distance
- central lymphoid organ (primary lymphoid organ)
Lymphoid organ in which lymphocytes develop. In adult mammals these are the thymus and bone marrow.
- central nervous system (CNS)
Main information-processing organ of the nervous system. In vertebrates it consists of the brain and spinal cord.
- centriole
Short cylindrical array of microtubules, closely similar in structure to a basal body. A pair of centrioles is usually found at the center of a centrosome in animal cells.
- centromere
Constricted region of a mitotic chromosome that holds sister chromatids together. It is also the site on the DNA where the kinetochore forms that captures microtubules from the mitotic spindle.
- centrosome cycle
Duplication of the centrosome (during interphase) and separation of the two new centrosomes (at the beginning of mitosis), which provides two centrosomes to form the poles of the mitotic spindle.
- centrosome
Centrally located organelle of animal cells that is the primary microtubule-organizing center and acts as the spindle pole during mitosis. In most animal cells it contains a pair of centrioles.
- CG island
Region of DNA with a greater than average density of CG sequences; these regions generally remain unmethylated.
- CGN
see cis Golgi network
- channel protein
Membrane transport protein that forms an aqueous pore in the membrane through which a specific solute, usually an ion, can pass.
- chaperone (molecular chaperone)
Protein that helps other proteins avoid misfolding pathways that produce inactive or aggregated polypeptides.
- checkpoint
Point in the eucaryotic cell-division cycle where progress through the cycle can be halted until conditions are suitable for the cell to proceed to the next stage.
- chelate
Combine reversibly, usually with high affinity, with a metal ion such as iron, calcium, or magnesium.
- chemical group
Set of covalently linked atoms, such as a hydroxyl group (–OH) or an amino group (–NH2), the chemical behavior of which is well characterized.
- chemiosmotic coupling
Mechanism in which a gradient of hydrogen ions (a pH gradient) across a membrane is used to drive an energy-requiring process, such as ATP production or the rotation of bacterial flagella.
- chemokine
Small secreted protein that attracts cells, such as white blood cells, to move towards its source. Important in the functioning of the immune system.
- chemotaxis
Directed movement of a cell or organism towards or away from a diffusible chemical.
- chiasma (chiasmata)
X-shaped connection visible between paired homologous chromosomes in division I of meiosis, and which represents a site of crossing-over.
- chlorophyll
Light-absorbing green pigment that plays a central part in photosynthesis in bacteria, plants, and algae.
- chloroplast
Organelle in green algae and plants that contains chlorophyll and carries out photosynthesis. It is a specialized form of plastid.
- cholesterol
Lipid molecule with a characteristic four-ring steroid structure that is an important component of the plasma membranes of animal cells. (See Figure 10–10.)
- chondrocyte (cartilage cell)
Connective-tissue cell that secretes the matrix of cartilage.
- chromaffin cell
Cell that stores adrenaline in secretory vesicles and secretes it in times of stress when stimulated by the nervous system.
- chromatid
One copy of a chromosome formed by DNA replication that is still joined at the centromere to the other copy. The two identical chromatids are called sister chromatids.
- chromatin
Complex of DNA, histones, and nonhistone proteins found in the nucleus of a eucaryotic cell. The material of which chromosomes are made.
- chromatography
Biochemical technique in which a mixture of substances is separated by charge, size, or some other property by allowing it to partition between a moving phase and a stationary phase. (See affinity chromatography, DNA affinity chromatography, high-performance liquid chromatography.)
- chromosomal crossing-over
The exchange of DNA between paired homologous chromosomes in division I of meiosis. It is a sign of genetic recombination and the crossovers (chiasmata) are visible in the light microscope. (See Figure 20–10.)
- chromosome
Structure composed of a very long DNA molecule and associated proteins that carries part (or all) of the hereditary information of an organism. Especially evident in plant and animal cells undergoing mitosis or meiosis, where each chromosome becomes condensed into a compact rodlike structure visible under the light microscope.
- chromosome condensation
Process by which a chromosome becomes packed up into a more compact structure prior to M phase of the cell cycle.
- cilium (cilia)
Hairlike extension of a eucaryotic cell containing a core bundle of microtubules and capable of performing repeated beating movements. Cilia are found in large numbers on the surface of many cells, and are responsible for the swimming of many single-celled organisms.
- circadian clock
Internal cyclical process that produces a particular change in a cell or organism with a period of around 24 hours, for example the sleep-wakefulness cycle in humans.
- cis face
Face of a Golgi stack at which material enters the organelle. It is adjacent to the cis Golgi network.
- cis Golgi network (CGN)
Network of interconnected cisternae and tubules which receives vesicles from the endoplasmic reticulum and transfers material to the cis face of the Golgi apparatus.
- cisterna (cisternae)
Flattened membrane-bounded compartment, as found in the endoplasmic reticulum or Golgi apparatus.
- citric acid cycle (tricarboxylic acid (TCA) cycle, Krebs cycle)
Central metabolic pathway found in aerobic organisms. Oxidizes acetyl groups derived from food molecules to CO2 and H2O. In eucaryotic cells it occurs in the mitochondria.
- CKI
- class I MHC molecule
One of the two classes of MHC molecule. It is present on almost all cell types and presents viral peptides on the surface of virus-infected cells, where they are recognized by cytotoxic T cells. (See Figure 24–49.)
- class II MHC molecule
One of the two classes of MHC molecule. It is present on “professional” antigen-presenting cells and presents foreign peptides to helper T cells. (See Figure 24–49.)
- class switching
The change from making one class of immunoglobulin (for example IgM) to making another class (for example IgG) that many B cells undergo during the course of an immune response.
- classical pathway
A pathway for activating the complement system that is initiated by IgG or IgM antibodies bound to the surface of a microbe.
- clathrin
Protein that assembles into a polyhedral cage on the cytosolic side of a membrane so as to form a clathrin-coated pit, which buds off by endocytosis to form an intracellular clathrin-coated vesicle.
- clathrin-coated pit
Region of plasma membrane of animal cells that is coated with the protein clathrin on its cytosolic face. Such regions are continually forming and budding off by endocytosis to form intracellular clathrin-coated vesicles containing extracellular fluid and the materials dissolved in it.
- cleavage
Physical splitting of a cell into two.
Specialized type of cell division seen in many early embryos whereby a large cell becomes subdivided into many smaller cells without growth.
- clonal selection theory
Theory that explains how the adaptive immune system can respond to millions of different antigens in a highly specific way. From a population of lymphocytes with a vast repertoire of randomly generated antigen specificities, a given foreign antigen activates (selects) only those cells with the corresponding antigen specificity.
- clone
Population of cells or organisms formed by repeated (asexual) division from a common cell or organism. Also used as a verb: “to clone a gene” means to produce many copies of a gene by repeated cycles of replication.
- cloning vector
A small DNA molecule, usually derived from a bacteriophage or plasmid, which is used to carry the fragment of DNA to be cloned into the recipient cell, and which enables the DNA fragment to be replicated.
- coated vesicle
Small membrane-bounded organelle with a cage of proteins (the coat) on its cytosolic surface. It is formed by the pinching off of a coated region of membrane (coated pit). Some coats are made of clathrin, whereas others are made from other proteins.
- codon
Sequence of three nucleotides in a DNA or messenger RNA molecule that represents the instruction for incorporation of a specific amino acid into a growing polypeptide chain.
- coenzyme
Small molecule tightly associated with an enzyme that participates in the reaction that the enzyme catalyzes, often by forming a covalent bond to the substrate. Examples include biotin, NAD+, and coenzyme A.
- coenzyme A
Small molecule used in the enzymatic transfer of acyl groups in the cell. (See also acetyl CoA.)
- cofactor
Inorganic ion or coenzyme that is required for an enzyme’s activity.
- cohesin, cohesin complex
Complex of proteins that holds sister chomatids together along their length before their separation.
- coiled-coil
Especially stable rodlike structure in proteins which is formed by two of these α helices coiled around each other.
- collagen fibril
Extracellular structure formed by self-assembly of secreted fibrillar collagen subunits. An abundant constituent of the extracellular matrix in many animal tissues.
- collagen
Fibrous protein rich in glycine and proline that is a major component of the extracellular matrix and connective tissues. Exists in many forms: type I, the most common, is found in skin, tendon, and bone; type II is found in cartilage; type IV is present in basal laminae.
- colony-stimulating factor (CSF)
General name for the numerous signal molecules that control the differentiation of blood cells.
- colorectal tumor
Common carcinoma of the epithelium lining the colon and rectum.
- combinatorial control
Describes the control of a step in a cellular process, such as the initiation of DNA transcription, by a combination of proteins rather than by any individual one.
- communicating junction
Type of cell junction that allows the passage of chemical or electrical signals from one cell to another.
- compartment
Regions in the embryo that are formed exclusively from the descendants of a few founder cells; there is no cell movement beween compartments once delimited.
- complement system
System of serum proteins activated by antibody–antigen complexes or by microorganisms. Helps eliminate pathogenic microorganisms by directly causing their lysis or by promoting their phagocytosis.
- complementary DNA
see cDNA
- complementary
Two nucleic acid sequences are said to be complementary if they can form a perfect base-paired double helix with each other.
- complex oligosaccharide
Chain of sugars attached to a glycoprotein that is generated by trimming of the original oligosaccharide attached in the endoplasmic reticulum and subsequent addition of further sugars. (See Figure 13–25.)
- complex
Assembly of molecules that are held together by noncovalent bonds. Protein complexes perform most cell functions.
- condensation reaction
Chemical reaction in which two molecules are covalently linked through –OH groups with the removal of a molecule of water.
- condensin, condensin complex
Complex of proteins involved in chromosome condensation prior to mitosis. Target for the M-Cdk.
- conditional mutation
A mutation that changes a protein or RNA molecule so that its function is altered only under some conditions, such as at an unusually high or an unusually low temperature.
- confocal microscope
Type of light microscope that produces a clear image of a given plane within a solid object. It uses a laser beam as a pinpoint source of illumination and scans across the plane to produce a two-dimensional ‘optical section.’
- conformation
The spatial arrangement of atoms in three dimensions in a macromolecule such as a protein or nucleic acid.
- connective tissue
Any supporting tissue that lies between other tissues and consists of cells embedded in a relatively large amount of extracellular matrix. Includes bone, cartilage, and loose connective tissue.
- connective-tissue cell
Any of the various cell types found in connective tissue, e.g. fibroblasts, cartilage cells (chondrocytes), bone cells (osteoblasts and osteocytes), fat cells (adipocytes) and smooth muscle cells.
- connexon
Water-filled pore in the plasma membrane formed by a ring of six protein subunits. Part of a gap junction: connexons from two adjoining cells join to form a continuous channel between the two cells.
- consensus sequence
Average or most typical form of a sequence that is reproduced with minor variations in a group of related DNA, RNA, or protein sequences. The consensus sequence shows the nucleotide or amino acid most often found at each position. The preservation of a consensus implies that the sequence is functionally important. (See Figure 6–12.)
- constitutive secretory pathway
Pathway present in all cells by which molecules such as plasma membrane proteins are continually delivered to the plasma membrane from the Golgi apparatus in vesicles that fuse with the plasma membrane. (See also default pathway.)
- constitutive
Produced in constant amount; opposite of regulated.
- contact-dependent signaling
Cell–cell communication in which the signal molecule remains bound to the signaling cell and only influences cells that physically contact it.
- contractile ring
Ring containing actin and myosin that forms under the surface of animal cells undergoing cell division and contracts to pinch the two daughter cells apart.
- convergent extension
Cellular rearrangement within a tissue that causes it to extend in one dimension (e.g. length) and shrink in another (e.g. width).
- cooperativity
Phenomenon in which the binding of one ligand molecule to a target molecule promotes the binding of successive ligand molecules. Seen in the assembly of large complexes, as well as in enzymes and receptors composed of multiple allosteric subunits, where it sharpens the response to a ligand. (See Figure 15–22.)
- cortical granule
Specialized secretory vesicle present under the plasma membrane of unfertilized eggs, including those of mammals; after fertilization it is involved in preventing the entry of further sperm.
- co-translational
Describes import of a protein into the endoplasmic reticulum before the polypeptide chain is completely synthesized.
- co-transport (coupled transport)
Membrane transport process in which the transfer of one molecule depends on the simultaneous or sequential transfer of a second molecule.
- coupled reaction
Linked pair of chemical reactions in which the free energy released by one of the reactions serves to drive the other.
- covalent bond
Stable chemical link between two atoms produced by sharing one or more pairs of electrons.
- crista (cristae)
One of the folds of the inner mitochondrial membrane.
A sensory structure in the inner ear.
- critical concentration
Concentration of a protein monomer, such as actin or tubulin, that is in equilibrium with the assembled form of the protein (i.e. assembled into actin filaments or microtubules respectively). (See Panel 16–2, pp. 912–913.)
- crossing-over
- cryoelectron microscopy
Electron microscopy technique in which the objects to be viewed, such as macromolecules and viruses, are rapidly frozen.
- cryptochrome
Flavoprotein responsive to blue light, found in both plants and animals. In animals it is involved in circadian rhythms.
- cut-and-paste transposition
Type of movement of a transposable element in which it is cut out of the DNA and inserted into a new site by a special transposase enzyme.
- cyclic AMP (cAMP)
Nucleotide that is generated from ATP by adenylyl cyclase in response to stimulation of many types of cell-surface receptors. cAMP acts as an intracellular signaling molecule by activating cyclic-AMP-dependent kinase (protein kinase A, PKA). It is hydrolyzed to AMP by a phosphodiesterase.
- cyclic AMP-dependent protein kinase (protein kinase A, PKA)
Enzyme that phosphorylates target proteins in response to a rise in intracellular cyclic AMP.
- cyclic GMP
Small soluble intracellular signaling molecule formed from GTP by the enzyme guanylyl cyclase in response to photoreceptor stimulation in the retina.
- cyclin
Protein that periodically rises and falls in concentration in step with the eucaryotic cell cycle. Cyclins activate crucial protein kinases (called a cyclin-dependent protein kinase, or Cdk) and thereby help control progression from one stage of the cell cycle to the next.
- cyclin-Cdk complex
Protein complexes that are formed periodically during the eucaryotic cell cycle as the level of cyclin increases, and in which the cyclin-dependent kinase (Cdk) becomes partially activated.
- cyclin-dependent kinase (Cdk)
Protein kinase that has to be complexed with a cyclin protein in order to act. Different Cdk-cyclin complexes trigger different steps in the cell-division cycle by phosphorylating specific target proteins.
- cytochrome b-c1 complex
Second of the three electron-driven proton pumps in the respiratory chain. It accepts electrons from ubiquinone.
- cytochrome oxidase complex
Third of the three electron-driven proton pumps in the respiratory chain. It accepts electrons from cytochrome c and generates water using molecular oxygen as an electron acceptor.
- cytochrome
Colored, heme-containing protein that transfers electrons during cellular respiration and photosynthesis.
- cytokine
Extracellular signal protein or peptide that acts as a local mediator in cell–cell communication.
- cytokine receptor
Type of cell-surface receptor whose ligands are cytokines such as interferons, growth hormone and prolactin, and which acts through the Jak-STAT pathway.
- cytokinesis
Division of the cytoplasm of a plant or animal cell into two, as distinct from the division of its nucleus (which is mitosis)
- cytoplasm
Contents of a cell that are contained within its plasma membrane but, in the case of eucaryotic cells, outside the nucleus.
- cytoskeleton
System of protein filaments in the cytoplasm of a eucaryotic cell that gives the cell shape and the capacity for directed movement. Its most abundant components are actin filaments, microtubules, and intermediate filaments.
- cytosol
Contents of the main compartment of the cytoplasm, excluding membrane-bounded organelles such as endoplasmic reticulum and mitochondria. Originally defined operationally as the cell fraction remaining after membranes, cytoskeletal components, and other organelles have been removed by low-speed centrifugation.
- cytotoxic T cell
Type of T cell responsible for killing infected cells.
- ΔG°
see standard free-energy change
- ΔG
see free-energy change
- dalton
Unit of molecular mass. Approximately equal to the mass of a hydrogen atom (1.66 × 10–24 g).
- default pathway
Constitutive secretory pathway that automatically delivers material from the Golgi apparatus to the plasma membrane if no other sorting signals are present.
- degenerate
Not a moral judgment but an adjective that describes multiple states that amount to the same thing: different triplet combinations of nucleotide bases (codons) that code for the same amino acid, for example.
- deletion
Type of mutation in which a single nucleotide or sequence of nucleotides has been removed from the DNA.
- denaturation
Dramatic change in conformation of a protein or nucleic acid caused by heating or by exposure to chemicals and usually resulting in the loss of biological function.
- dendrite
Extension of a nerve cell, typically branched and relatively short, that receives stimuli from other nerve cells.
- dendritic cell
Cell derived from bone marrow and present in lymphoid and other tissues that is specialized for the uptake of particulate material by phagocytosis and which acts as a “professional” antigen-presenting cell in immune responses.
- deoxyribonucleic acid
see DNA
- desensitization
see adaptation
- desmosome
Type of anchoring cell–cell junction, usually formed between two epithelial cells, characterized by dense plaques of protein into which intermediate filaments in the two adjoining cells insert.
- detergent
Type of small amphipathic molecule that tends to coalesce in water, with its hydrophobic tails buried and its hydrophilic heads exposed. It is widely used to solubilize membrane proteins.
- determined
In developmental biology, an embryonic cell is said to be determined if it has become committed to a particular specialized path of development. This determination reflects a change in the internal character of the cell, and it precedes the much more readily detected process of cell differentiation.
- development
Succession of changes that take place in an organism as a fertilized egg gives rise to an adult plant or animal.
- diacylglycerol
Lipid produced by the cleavage of inositol phospholipids in response to extracellular signals. Composed of two fatty acid chains linked to glycerol, it serves as a signaling molecule to help activate protein kinase C.
- dideoxy method
The standard method of DNA sequencing.
- differentiation
Process by which a cell undergoes a change to an overtly specialized cell type.
- diffraction pattern
Pattern set up by wave interference between radiation transmitted or scattered by different parts of an object.
- diffusion
Net drift of molecules in the direction of lower concentration due to random thermal movement.
- diploid
Containing two sets of homologous chromosomes and hence two copies of each gene or genetic locus.
- diplotene
Fourth stage of division I of meiosis, in which chiasmata are first seen.
- disaccharide
Carbohydrate molecule consisting of two covalently joined monosaccharide units. (See Panel 2–4, p. 116–117.)
- dissociation constant (Kd)
Measure of the tendency of a complex to dissociate. For components A and B and the binding equilibrium A + B ⇌ AB, the dissociation constant is given by [A][B]/[AB], and it is smaller the tighter the binding between A and B. (See also association constant.)
- disulfide bond (SS)
Covalent linkage formed between two sulfhydryl groups on cysteines. For extracellular proteins, a common way of joining two proteins together or linking different parts of the same protein. Formed in the endoplasmic reticulum of eucaryotic cells.
- division I of meiosis
The first cell division of meiosis, in which the members of each pair of (duplicated) homologous chromosomes are segregated to opposite poles of the dividing cell.
- division II of meiosis
The second cell division of meiosis, in which the chromatids of each duplicated chromosome are segregated to opposite poles of the dividing cell.
- DNA (deoxyribonucleic acid)
Polynucleotide formed from covalently linked deoxyribonucleotide units. It serves as the store of hereditary information within a cell and the carrier of this information from generation to generation.
- DNA affinity chromatography
Technique for purifying sequence-specific DNA-binding proteins by their binding to a matrix to which the appropriate DNA fragments are attached.
- DNA footprinting
Technique for determining the DNA sequence to which a DNA-binding protein binds.
- DNA helicase
Enzyme that is involved in opening the DNA helix into its single strands for DNA replication.
- DNA library
Collection of cloned DNA molecules, representing either an entire genome (genomic library) or DNA copies of the messenger RNA produced by a cell (cDNA library).
- DNA ligase
Enzyme that joins the ends of two strands of DNA together with a covalent bond to make a continuous DNA strand.
- DNA methylation
Addition of a methyl group to DNA. Extensive methylation of the cytosine base in CG sequences is used in vertebrates to keep genes in an inactive state.
- DNA microarray
Technique for analyzing the simultaneous expression of large numbers of genes in cells, in which isolated cellular RNA is hybridized to a large array of short DNA probes immobilized on glass slides.
- DNA polymerase
Enzyme that synthesizes DNA by joining nucleotides together using a DNA template as a guide.
- DNA primase
Enzyme that synthesizes a short strand of RNA on a DNA template, producing a primer for DNA synthesis.
- DNA repair
Collective name for those biochemical processes that correct accidental changes in the DNA.
- DNA sequencing
Determination of the order of nucleotides in a DNA molecule. (See Figure 8–36.)
- DNA supercoiling
Additional twisting of the DNA helix that occurs in response to the superhelical tension created when, for example, a circular DNA is partly unwound (See Figure 6–20.)
- DNA topoisomerase
Enzyme that binds to DNA and reversibly breaks a phosphodiester bond in one or both strands, allowing the DNA to rotate at that point. It prevents DNA tangling during replication.
- DNA transcription
see transcription
- DNA tumor virus
A general term for a variety of different DNA viruses that can cause tumors.
- DNA-only transposon
Type of transposable element that exists as DNA throughout its life cycle. Many types move by cut-and-paste transposition.
- domain
see protein domain
- dominant negative mutation
Mutation that dominantly affects the phenotype by means of a defective protein or RNA molecule that interferes with the function of the normal gene product in the same cell.
- dominant
In genetics, refers to the member of a pair of alleles that is expressed in the phenotype of the organism while the other allele is not, even though both alleles are present. Opposite of recessive.
- dorsal
Relating to the back of an animal. Also the upper surface of a leaf, wing, etc.
- dorsoventral
Describes the axis running from the back to the belly of an animal or from the upper side to the underside of a structure.
- double helix
The three-dimensional structure of DNA, in which two DNA chains held together by hydrogen bonding between the bases are wound into a helix.
- Drosophila melanogaster
Species of small fly, commonly called a fruit fly, much used in genetic studies of development.
- dynamic instability
The property of sudden conversion from growth to shrinkage, and vice versa, in a protein filament such as a microtubule or actin filament. (See Panel 16–2, pp. 912–913.)
- dynamin
Cytosolic GTPase that binds to the neck of a clathrin-coated vesicle in the process of budding from the membrane, and which is involved in completing vesicle formation.
- dynein
Member of a family of large motor proteins that undergo ATP-dependent movement along microtubules. In cilia, dynein forms the side arms in the axoneme that cause adjacent microtubule doublets to slide past one another.
- dysplasia
A change in cell growth and behavior in a tissue in which the structure becomes disordered.
- ectoderm
Embryonic tissue that is the precursor of the epidermis and nervous system.
- effector cell
A cell that carries out the final response or function of a particular process. The main effector cells of the immune system, for example, are activated lymphocytes and phagocytes—the cells involved in destroying pathogens and removing them from the body.
- egg
The mature female gamete in sexually reproducing organisms. It is usually a large and immobile cell.
- elastin
Hydrophobic protein that forms extracellular extensible fibres (elastic fibres) that give tissues their stretchability and resilience.
- electrochemical gradient
The combined influence of a difference in the concentration of an ion on the two sides of the membrane and the electrical charge difference across the membrane (membrane potential). It produces a driving force that causes the ion to move across the membrane.
- electrochemical proton gradient
The result of a combined pH gradient (proton gradient) and the membrane potential.
- electron
Negatively charged subatomic particle that generally occupies orbitals surrounding the nucleus in an atom.
- electron acceptor
Atom or molecule that takes up electrons readily, thereby gaining an electron and becoming reduced.
- electron carrier
Molecule such as cytochrome c, which transfers an electron from a donor molecule to an acceptor molecule.
- electron donor
Molecule that easily gives up an electron, becoming oxidized in the process.
- electron microscope
Type of microscope that uses a beam of electrons to create the image.
- electron-transport chain
Series of electron carrier molecules along which electrons move from a higher to a lower energy level to a final acceptor molecule. The energy released during electron movement can be used to power various processes. Electron-transport chains present in the inner mitochondrial membrane and in the thylakoid membrane of chloroplasts generate a proton gradient across the membrane that is used to drive ATP synthesis.
- elongation factor
Protein required for the addition of amino acids to growing polypeptide chains on ribosomes.
- embryogenesis
- embryonic stem cell (ES cell)
Cell derived from the inner cell mass of the early mammalian embryo that can give rise to all the cells in the body. It can be grown in culture, genetically modified and inserted into a blastocyst to develop a transgenic animal.
- endocrine cell
Specialized animal cell that secretes a hormone into the blood. Usually part of a gland, such as the thyroid or pituitary gland.
- endocytic-exocytic cycle
The processes of endocytosis and exocytosis that, respectively, add and remove plasma membrane from the cell, resulting in no overall change in the cell’s surface area and volume.
- endocytosis
Uptake of material into a cell by an invagination of the plasma membrane and its internalization in a membrane-bounded vesicle. (See also pinocytosis and phagocytosis.)
- endoderm
Embryonic tissue that is the precursor of the gut and associated organs.
- endoplasmic reticulum (ER)
Labyrinthine membrane-bounded compartment in the cytoplasm of eucaryotic cells, where lipids are synthesized and membrane-bound proteins and secretory proteins are made.
- endosome
Membrane-bounded organelle in animal cells that carries materials newly ingested by endocytosis and passes many of them on to lysosomes for degradation.
- endothelial cell
Flattened cell type that forms a sheet (the endothelium) lining all blood vessels.
- enhancer
Regulatory DNA sequence to which gene regulatory proteins bind, influencing the rate of transcription of a structural gene that can be many thousands of base pairs away.
- entropy
Thermodynamic quantity that measures the degree of disorder in a system; the higher the entropy, the greater the disorder.
- enveloped virus
Virus with a capsid surrounded by a lipid membrane (the envelope), which is derived from the host cell plasma membrane when the virus buds from the cell.
- enzyme
Protein that catalyzes a specific chemical reaction.
- enzyme-linked receptor
Major type of cell-surface receptor in which the cytoplasmic domain either has enzymatic activity itself or is associated with an intracellular enzyme. In both cases enzymatic activity is stimulated by ligand binding to the receptor.
- epidermis
Epithelial layer covering the outer surface of the body. It has different structures in different animal groups. The outer layer of plant tissue is also called the epidermis.
- epimerization
Reaction that alters the steric arrangement around one atom, as in a sugar molecule.
- epinephrine
see adrenaline
- epithelial tissue
see epithelium
- epithelium (epithelia)
Coherent cell sheet formed from one or more layers of cells covering an external surface or lining a cavity.
- epitope
see antigenic determinant
- equilibrium constant (K)
Ratio of forward and reverse rate constants for a reaction and equal to the association constant. (See Figure 3–44.)
- equilibrium
State where there is no net change in a system. For example, equilibrium is reached in a chemical reaction when the forward and reverse rates are equal.
- ER lumen
The space enclosed by the membrane of the endoplasmic reticulum (ER).
- ER resident protein
Protein that remains in the endoplasmic reticulum (ER) or its membranes and carries out its function there, as opposed to proteins that are present in the ER only in transit.
- ER retention signal
Short amino acid sequence on a protein that prevents it moving out of the endoplasmic reticulum (ER). Found on proteins that are resident in the ER and function there.
- ER signal sequence
N-terminal signal sequence that directs proteins to enter the endoplasmic reticulum (ER). It is cleaved off by signal peptidase after entry.
- ER
- erythrocyte (red blood cell)
Small, hemoglobin-containing blood cell of vertebrates that transports oxygen and carbon dioxide to and from tissues.
- erythropoietin
Growth factor that stimulates the production of red blood cells. It is produced by the kidney and acts on precursor cells in bone marrow.
- ES cell
see embryonic stem cell
- Escherichia coli (E. coli)
Rodlike bacterium normally found in the colon of humans and other mammals and widely used in biomedical research.
- ester
Molecule formed by the condensation reaction of an alcohol group with an acidic group. Phosphate groups usually form esters when linked to a second molecule. (See Panel 2–1, 110–111.)
- ethyl (CH2CH3)
Hydrophobic chemical group derived from ethane (CH3CH3).
- eucaryote (eukaryote)
Organism composed of one or more cells with a distinct nucleus and cytoplasm. Includes all forms of life except viruses and procaryotes (bacteria and archea).
- euchromatin
Region of an interphase chromosome that stains diffusely; “normal” chromatin, as opposed to the more condensed heterochromatin.
- exocytosis
Process by which most molecules are secreted from a eucaryotic cell. These molecules are packaged in membrane-bounded vesicles that fuse with the plasma membrane, releasing their contents to the outside.
- exon
Segment of a eucaryotic gene that consists of a sequence of nucleotides that will be represented in messenger RNA or the final transfer RNA or ribosomal RNA. In protein-coding genes, exons encode amino acids in the protein. An exon is usually adjacent to a noncoding DNA segment called an intron.
- expression vector
A virus or plasmid that carries a DNA sequence into a suitable host cell and there directs the synthesis of the protein encoded by the sequence.
- expression
Production of an observable phenotype by a gene—usually by directing the synthesis of a protein.
- extracellular matrix
Complex network of polysaccharides (such as glycosaminoglycans or cellulose) and proteins (such as collagen) secreted by cells. Serves as a structural element in tissues and also influences their development and physiology.
- facilitated diffusion
- FADH2 (reduced flavin adenine dinucleotide)
Activated carrier molecule that is produced by the citric acid cycle.
- FAK
- Fas protein (Fas)
Membrane-bound receptor that initiates apoptosis in the receptor-bearing cell after binding to its ligand (Fas ligand).
- fat
Energy-storage lipid in cells. It is composed of triglycerides—fatty acids esterified with glycerol.
- fat cell
Connective-tissue cell that produces and stores fat in animals.
- fatty acid
Compound such as palmitic acid that has a carboxylic acid attached to a long hydrocarbon chain. Used as a major source of energy during metabolism and as a starting point for the synthesis of phospholipids. (See Panel 2–5, pp. 118–119.)
- Fc receptor
One of a family of receptors specific for the invariant constant region (Fc region) of immunoglobulins (other than IgM and IgD); different Fc receptors are specific for IgG, IgA, IgE and their subclasses.
- feedback inhibition
Type of regulation of metabolism in which an enzyme acting early in a reaction pathway is inhibited by a late product of that pathway.
- fermentation
Anaerobic energy-yielding metabolic pathway in which pyruvate produced by glycolysis is converted, for example, into lactate or ethanol, with the conversion of NADH to NAD+.
- fertilization
Fusion of a male and a female gamete (both haploid) to form a diploid zygote, which develops into a new individual.
- fibrillar collagen
Type of collagen molecule which assembles into rope-like structures. Collagens type I (common in skin), II, III, V and XI are of this type.
- fibroblast
Common cell type found in connective tissue. Secretes an extracellular matrix rich in collagen and other extracellular matrix macromolecules. Migrates and proliferates readily in wounded tissue and in tissue culture.
- fibronectin
Extracellular matrix protein that is involved in adhesion of cells to the matrix and the guidance of migrating cells during embryogenesis. Integrins on the cell surface are receptors for fibronectin.
- filopodium (filopodia)
Thin, spike-like protrusion with an actin filament core, generated on the leading edge of a crawling animal cell.
- fission yeast
Common name often given to the yeast Schizosaccharomyces pombe, a common experimental organism. It divides to give two equal-sized cells.
- fixative
Chemical reagent such as formaldehyde or osmium tetroxide used to preserve cells for microscropy. Samples treated with these reagents are said to be “fixed,” and the process is called fixation.
- flagellum (flagella)
Long, whiplike protrusion whose undulations drive a cell through a fluid medium. Eucaryotic flagella are longer versions of cilia. Bacterial flagella are smaller and completely different in construction and mechanism of action.
- fluid-phase endocytosis
Type of endocytosis in which small vesicles bud off internally from the plasma membrane, carrying extracellular fluid and dissolved material into the cell. (See also pinocytosis.)
- fluorescein
Fluorescent dye that fluoresces green when illuminated with blue light or ultraviolet light.
- fluorescence microscope
Microscope designed to view material stained with fluorescent dyes. Similar to a light microscope but the illuminating light is passed through one set of filters before the specimen, to select those wavelengths that excite the dye, and through another set of filters before it reaches the eye, to select only those wavelengths emitted when the dye fluoresces.
- fluorescent dye
Molecule that absorbs light at one wavelength and responds by emitting light at another wavelength. The emitted light is of longer wavelength (and hence of lower energy) than the light absorbed.
- fluorescent resonance energy transfer (FRET)
Technique for monitoring the closeness of two fluorescently labeled molecules (and thus their interaction) in cells.
- focal adhesion kinase (FAK)
Cytoplasmic tyrosine kinase present at cell-matrix junctions (focal adhesions) in association with the cytoplasmic tails of integrins.
- focal adhesion, focal contact (adhesion plaque)
A type of anchoring cell junction, forming a small region on the surface of a fibroblast or other cell that is anchored to the extracellular matrix. Attachment is mediated by transmembrane proteins such as integrins, which are linked, through other proteins, to actin filaments in the cytoplasm.
- follicle cell
One of the cell types that surround a developing oocyte or egg.
- free energy (G)
The energy that can be extracted from a system to drive reactions. Takes into account changes in both energy and entropy.
- free ribosome
Ribosome that is free in the cytosol, unattached to any membrane. It is the site of synthesis of all proteins encoded by the nuclear genome other than those destined to enter the endoplasmic reticulum.
- free-energy change (G)
Change in the free energy during a reaction: the free energy of the product molecules minus the free energy of the starting molecules. A large negative value of ΔG indicates that the reaction has a strong tendency to occur. (See Panel 2–7, pp. 122–123.)
- freeze-fracture electron microscopy
Technique for studying membrane structure, in which the membrane of a frozen cell is fractured along the interior of the bilayer, separating it into the two monolayers with the interior faces exposed.
- FRET
- fungus (fungi)
Kingdom of eucaryotic organisms that includes the yeasts, molds, and mushrooms. Many plant diseases and a relatively small number of animal diseases are caused by fungi.
- γ-tubulin ring complex (TuRC)
Protein complex containing γ-tubulin and other proteins that is an efficient nucleator of microtubules.
- G
see free energy
- G0
G-“zero” phase. State of withdrawal from the eucaroytic cell-division cycle by entry into a quiescent G1 phase. A common state for differentiated cells.
- G1 phase
Gap 1 phase of the eucaryotic cell-division cycle, between the end of cytokinesis and the start of DNA synthesis.
- G1/S-Cdk
Complex formed in vertebrate cells by a G1/S-cyclin and the corresponding cyclin-dependent kinase (Cdk).
- G1-Cdk
Complex formed in vertebrate cells by a G1-cyclin and the corresponding cyclin-dependent kinase (Cdk).
- G2 phase
Gap 2 phase of the eucaryotic cell-division cycle, between the end of DNA synthesis and the beginning of mitosis.
- GAG
see glycosaminoglycan
- gamete
Specialized haploid cell, either a sperm or an egg, serving for sexual reproduction.
- ganglion (ganglia)
Cluster of nerve cells and associated glial cells located outside the central nervous system.
- ganglioside
Any glycolipid having one or more sialic acid residues in its structure. Found in the plasma membrane of eucaryotic cells and especially abundant in nerve cells.
- gap junction
Communicating cell–cell junction that allows ions and small molecules to pass from the cytoplasm of one cell to the cytoplasm of the next.
- gastrulation
The stage in animal embryogenesis during which the embryo is transformed from a ball of cells to a structure with a gut (a gastrula).
- gene activator protein
A gene regulatory protein that when bound to its regulatory sequence in DNA activates transcription.
- gene control region
DNA sequences required to initiate transcription of a given gene and control the rate of initiation.
- gene conversion
Process by which DNA sequence information can be transferred from one DNA helix (which remains unchanged) to another DNA helix whose sequence is altered. It occurs occasionally during general recombination.
- gene regulatory protein
General name for any protein that binds to a specific DNA sequence to alter the expression of a gene.
- gene repressor protein
A gene regulatory protein that prevents the initiation of transcription.
- gene
Region of DNA that controls a discrete hereditary characteristic, usually corresponding to a single protein or RNA. This definition includes the entire functional unit, encompassing coding DNA sequences, noncoding regulatory DNA sequences, and introns.
- general recombination, general genetic recombination
Recombination that takes place between two homologous chromosomes (as in meiosis).
- general transcription factor
Any of the proteins whose assembly around the TATA box is required for the initiation of transcription of most eucaryotic genes.
- genetic code
Set of rules specifying the correspondence between nucleotide triplets (codons) in DNA or RNA and amino acids in proteins.
- genetic map
Map of the chromosomes in which the distance of genes relative to each other is determined by the amount of genetic recombination that occurs between them.
- genetic recombination
see recombination
- genetic screen
A search through a large collection of mutants for a mutant with a particular phenotype.
- genome
The totality of genetic information belonging to a cell or an organism; in particular, the DNA that carries this information.
- genomic DNA
DNA constituting the genome of a cell or an organism. Often used in contrast to cDNA (DNA prepared by reverse transcription from messenger RNA). Genomic DNA clones represent DNA cloned directly from chromosomal DNA, and a collection of such clones from a given genome is a genomic DNA library.
- genomic imprinting
Situation where a gene is either expressed or not expressed in the embryo depending on which parent it is inherited from.
- genomics
The science of studying the DNA sequences and properties of entire genomes.
- genotype
Genetic constitution of an individual cell or organism.
- germ cell
Precursor cell that will give rise to gametes.
- germ line
The lineage of germ cells (which contribute to the formation of a new generation of organisms), as distinct from somatic cells (which form the body and leave no descendants).
- GFP
- giga-
Prefix denoting 109. (From Greek gigas, giant.)
- Gi
- glial cell
Supporting cell of the nervous system, including oligodendrocytes and astrocytes in the vertebrate central nervous system and Schwann cells in the peripheral nervous system.
- globular protein
Any protein with an approximately rounded shape. Such proteins are contrasted with highly elongated, fibrous proteins such as collagen.
- glucose
Six-carbon sugar that plays a major role in the metabolism of living cells. Stored in polymeric form as glycogen in animal cells and as starch in plant cells. (See Panel 2–4, pp. 116–117.)
- glutaraldehyde
Small reactive molecule with two aldehyde groups that is often used as a cross-linking fixative.
- glycerol
Small organic molecule that is the parent compound of many small molecules in the cell, including phospholipids.
- glycocalyx (cell coat)
Carbohydrate-rich layer that forms the outer coat of a eucaryotic cell. Composed of the oligosaccharides linked to intrinsic plasma membrane glycoproteins and glycolipids, as well as glycoproteins and proteoglycans that have been secreted and reabsorbed onto the cell surface.
- glycogen
Polysaccharide composed exclusively of glucose units used to store energy in animal cells. Large granules of glycogen are especially abundant in liver and muscle cells.
- glycolipid
Membrane lipid molecule with a sugar residue or oligosaccharide attached to the polar headgroup. (See Panel 2–5, pp. 118–119.)
- glycolysis
Ubiquitous metabolic pathway in the cytosol in which sugars are incompletely degraded with production of ATP. (Literally, “sugar splitting.”)
- glycoprotein
Any protein with one or more oligosaccharide chains covalently linked to amino-acid side chains. Most secreted proteins and most proteins exposed on the outer surface of the plasma membrane are glycoproteins.
- glycosaminoglycan (GAG)
Long, linear, highly charged polysaccharide composed of a repeating pair of sugars, one of which is always an amino sugar. Mainly found covalently linked to a protein core in extracellular matrix proteoglycans. Examples include chondroitin sulfate, hyaluronic acid, and heparin.
- glycosylation
The process of adding one or more sugars to a protein or lipid molecule. (See also O-linked glycosylation, N-linked glycosylation.)
- glycosylphosphatidylinositol anchor (GPI anchor)
Type of lipid linkage by which some membrane proteins are bound to the membrane. It is formed as the proteins travel through the endoplasmic reticulum.
- Golgi apparatus (Golgi complex)
Membrane-bounded organelle in eucaryotic cells in which proteins and lipids transferred from the endoplasmic reticulum are modified and sorted. It is the site of synthesis of many cell wall polysaccharides in plants and extracellular matrix glycosaminoglycans in animal cells.
- GPI anchor
see glycosylphosphatidylinositol anchor
- G-protein
see GTP-binding protein
- G-protein-linked receptor
Cell-surface receptor that associates with an intracellular trimeric GTP-binding protein (G protein) after receptor activation by an extracellular ligand. These receptors are seven-pass transmembrane proteins.
- Gq
Class of receptor-coupled G protein that activates phospholipase C-β and originates the inositol phospholipid signaling pathway.
- grana (singular granum)
Stacked membrane discs (thylakoids) in chloroplasts that contain chlorophyll and are the site of the light-trapping reactions of photosynthesis.
- granulocyte
Category of white blood cell distinguished by conspicuous cytoplasmic granules. Includes neutrophils, basophils, and eosinophils.
- gray crescent
Band of pale pigmentation that appears in the egg of some species of amphibian opposite the site of sperm entry following fertilization. Caused by rotation of the egg cortex and associated pigment granules. Marks the future dorsal side.
- green fluorescent protein (GFP)
Fluorescent protein isolated from a jellyfish. Widely used as a marker in cell biology.
- growth cone
Migrating motile tip of a growing nerve cell axon or dendrite.
- growth factor
Extracellular polypeptide signal molecule that can stimulate a cell to grow or proliferate. Examples are epidermal growth factor (EGF) and platelet-derived growth factor (PDGF). Most growth factors also have other actions.
- growth regulator
- Gs
- GTP (guanosine 5-triphosphate)
Nucleoside triphosphate produced by phosphorylating GDP (guanosine diphosphate). Like ATP it releases a large amount of free energy on hydrolysis of its terminal phosphate group. It has a special role in microtubule assembly, protein synthesis, and cell signaling.
- GTPase
Enzyme activity that converts GTP to GDP. Also the common name used for monomeric GTP-binding proteins. (See GTP-binding protein.)
- GTPase-activating protein (GAP)
Protein that binds to a GTP-binding protein and inactivates it by stimulating its GTPase activity so that it hydrolyzes its bound GTP to GDP.
- GTP-binding protein, G protein
Protein with GTPase activity that binds GTP, which activates the protein. The intrinsic GTPase activity eventually converts the GTP to GDP which inactivates the protein. These GTPases act as molecular switches in, for example, intracellular signaling pathways. One family is composed of three different subunits (heterotrimeric GTP-binding proteins). The members of the other, very large family are monomeric GTP-binding proteins; these are commonly referred to as monomeric GTPases.
- guanine nucleotide exchange factor (GEF)
Protein that binds to a GTP-binding protein and activates it by stimulating it to release its tightly bound GDP, thereby allowing it to bind GTP in its place.
- H+
see proton
- haploid
Having only one set of chromosomes, as in a sperm cell or a bacterium, as distinct from diploid (having two sets of chromosomes).
- heat shock protein (stress-response protein)
Protein synthesized in increased amounts in response to an elevated temperature or other stressful treatment, and which usually helps the cell to survive the stress. Prominent examples are hsp60 and hsp70.
- heavy chain (H chain)
The larger of the two types of polypeptide in an immunoglobulin molecule.
- HeLa cell
Line of human epithelial cells that grows vigorously in culture. Derived from a human cervical carcinoma.
- helix-loop-helix (HLH)
DNA-binding structural motif present in many gene regulatory proteins. Should not be confused with the helix-turn-helix.
- helper T cell
Type of T cell that helps stimulate B cells to make antibodies and activates macrophages to kill ingested microorganisms.
- heme
Cyclic organic molecule containing an iron atom that carries oxygen in hemoglobin and carries an electron in cytochromes. (See Figure 14–22.)
- hemidesmosome
Specialized anchoring cell junction between an epithelial cell and the underlying basal lamina.
- hemoglobin
The major protein in red blood cells that associates with O2 in the lungs by means of a bound heme group.
- hemopoiesis
Generation of blood cells, mainly in the bone marrow.
- hepatocyte
Liver cell.
- heterocaryon
Cell with two or more genetically different nuclei; produced by the fusion of two or more different cells.
- heterochromatin
Region of a chromosome that remains unusually condensed chromatin; transcriptionally inactive during interphase.
- heterodimer
Protein complex composed of two different polypeptide chains.
- heterozygote
Diploid cell or individual having two different alleles of one or more specified genes.
- high-energy bond
Covalent bond whose hydrolysis releases an unusually large amount of free energy under the conditions existing in a cell. A group linked to a molecule by such a bond is readily transferred from one molecule to another. Examples include the phosphodiester bonds in ATP and the thioester linkage in acetyl CoA.
- high-mannose oligosaccharide
Chain of sugars attached to a glycoprotein which contains many mannose residues. It is generated by a trimming of the original mannose-rich oligosaccharide that leaves most of the mannose residues with no subsequent addition of further sugars. (See Figure 13–26.)
- high-performance liquid chromatography (HPLC)
Type of chromatography that uses columns packed with tiny beads of matrix; the solution to be separated is pushed through under high pressure.
- histidine-kinase-associated receptor
Type of transmembrane receptor found in the plasma membrane of bacteria, yeast and plant cells, and involved, for example, in sensing stimuli that cause bacterial chemotaxis. Associated with a histidine protein kinase on its cytoplasmic side.
- histone
One of a group of small abundant proteins, rich in arginine and lysine, four of which form the nucleosome on the DNA in eucaryotic chromosomes.
- HIV
Human immunodeficiency virus, the retrovirus that is the cause of AIDS.
- HLH
see helix-loop-helix
- hnRNP protein (heterogeneous nuclear ribonuclear protein)
Any of a group of proteins that assemble on newly synthesized RNA, organizing it into a more compact form.
- Holliday junction
X-shaped structure observed in DNA undergoing recombination, in which the two DNA molecules are held together at the site of crossing-over, also called a cross-strand exchange.
- homeobox
Short (180 base pairs long) conserved DNA sequence that encodes a DNA-binding protein motif (homeodomain) famous for its presence in genes that are involved in orchestrating development in a wide range of organisms.
- homeodomain
DNA-binding domain that defines a class of gene regulatory proteins important in animal development.
- homeotic mutation
Mutation that causes cells in one region of the body to behave as though they were located in another, causing a bizarre disturbance of the body plan.
- homolog
One of two or more genes that are similar in sequence as a result of derivation from the same ancestral gene. The term covers both orthologs and paralogs.
See homologous chromosome.
- homologous
Describes organs or molecules that are similar because of their common evolutionary origin. Specifically it describes similarities in protein or nucleic acid sequence.
- homologous chromosome (homolog)
One of the two copies of a particular chromosome in a diploid cell, each copy being derived from a different parent.
- homozygote
Diploid cell or organism having two identical alleles of a specified gene or set of genes.
- hormone
Signal molecule secreted by an endocrine cell into the bloodstream, which can then carry it to distant target cells.
- housekeeping gene
Gene serving a function required in all the cell types of an organism, regardless of their specialized role.
- Hox complex
Two tightly linked clusters of genes in Drosophila (the bithorax and Antennapedia complexes) that control the differences between the different segments of the body. Homologous Hox complexes are found in other animals, where they also determine pattern along the anteroposterior axis.
- HPLC
- hybridization
In molecular biology, the process whereby two complementary nucleic acid strands form a double helix. Forms the basis of a powerful technique for detecting specific nucleotide sequences.
- hybridoma
Cell line used in the production of monoclonal antibodies. Obtained by fusing antibody-secreting B cells with cells of a lymphocyte tumor.
- hydrocarbon
Compound that has only carbon and hydrogen atoms. (See Panel 2–1, p 110–111.)
- hydrogen bond
Noncovalent bond in which an electropositive hydrogen atom is partially shared by two electronegative atoms.
- hydrolysis (adjective hydrolytic)
Cleavage of a covalent bond with accompanying addition of water, –H being added to one product of the cleavage and –OH to the other.
- hydronium ion (H3O)
- hydrophilic
Describes a polar molecule or part of a molecule that forms enough energetically favorable interactions with water molecules to dissolve readily in water. (Literally, “water loving.”)
- hydrophobic (lipophilic)
Describes a nonpolar molecule or part of a molecule that cannot form energetically favorable interactions with water molecules and therefore does not dissolve in water. (Literally, “water hating.”)
- hydrophobic force
Force exerted by the hydrogen-bonded network of water molecules that brings two nonpolar surfaces together by excluding water between them.
- hydroxyl (OH)
Chemical group consisting of a hydrogen atom linked to an oxygen, as in an alcohol.
- hypertonic
Describes any medium with a sufficiently high concentration of solutes to cause water to move out of a cell due to osmosis.
- hypervariable region
Any of three small regions within the variable region of an immunoglobulin light or heavy chain that show the highest variability from molecule to molecule. These regions determine the specificity of the antigen-binding site.
- hypotonic
Describes any medium with a sufficiently low concentration of solutes to cause water to move into a cell due to osmosis.
- IAP family
- Ig
see immunoglobulin
- Ig superfamily
Large family of proteins that contain immunoglobulin domains or immunoglobulin-like domains. Most are involved in cell-cell interactions or antigen recognition.
- image processing
Computer treatment of images gained from microscopy that reveal information not immediately visible to the eye.
- imaginal disc
Group of cells that are set aside in the Drosophila embryo and which will develop into an adult structure, e.g. eye, leg, wing.
- immature secretory vesicle
Secretory vesicle that appears to have just pinched off the Golgi stack. Its structure resembles that of a cisterna of the trans Golgi network.
- immortalization
Production of a cell line capable of an unlimited number of cell divisions. Can be the result of a chemical or viral transformation or of fusion of the original cells with cells of a tumor line.
- immune response
Response made by the immune system when a foreign substance or microorganism enters its body. (See also innate immune response, adaptive immune response, primary immune response, secondary immune response.)
- immune system
Population of lymphocytes and other white blood cells in the vertebrate body that defends it against infection.
- immunoglobulin (Ig)
An antibody molecule. Higher vertebrates have five classes of immunoglobulin—IgA, IgD, IgE, IgG, and IgM—each with a different role in the immune response.
- immunoglobulin domain (Ig domain)
Characteristic protein domain of about 100 amino acids that is found in immunoglobulin light and heavy chains. Similar domains, known as immunoglobulin-like (Ig-like) domains, are present in many other proteins involved in cell–cell interactions and antigen recognition and define the Ig superfamily.
- immunogold electron microscopy
Electron microscopy technique in which cellular structures or molecules of interest are labeled with antibodies tagged with electron-dense gold particles. These show up as black spots on the image.
- immunological memory
Long-lived state that follows a primary immune response to many antigens, in which subsequent encounter with that antigen will provoke a rapid secondary immune response.
- immunoprecipitation
Use of a specific antibody to draw the corresponding protein antigen out of solution. The technique can identify complexes of interacting proteins in cell extracts by using an antibody specific for one of the proteins to precipitate the complex.
- in situ hybridization
Technique in which a single-stranded RNA or DNA probe is used to locate a gene or a messenger RNA molecule in a cell or tissue by hybridization.
- in vitro
Term used by biochemists to describe a process taking place in an isolated cell-free extract. Also used by cell biologists to refer to cells growing in culture (in vitro), as opposed to in an organism (in vivo). (Latin for “in glass.”)
- in vivo
In an intact cell or organism. (Latin for “in life.”)
- induction
In developmental biology, a change in the developmental fate of one tissue caused by an interaction with another tissue. Such an interaction is called an inductive interaction.
- inflammatory response
Local response of a tissue to injury or infection—characterized by tissue redness, swelling, heat, and pain. Caused by invasion of white blood cells, which release various local mediators such as histamine.
- inhibitor of apoptosis family
see IAP family
- inhibitory G protein (Gi)
G protein that can regulate ion channels and inhibit the enzyme adenylyl cyclase.
- inhibitory neurotransmitter
Neurotransmitter that opens transmitter-gated Cl– or K+ channels in the postsynaptic membrane of a nerve or muscle cell and thus tends to inhibit the generation of an action potential.
- initiation factor
Protein that promotes the proper association of ribosomes with messenger RNA and is required for the initiation of protein synthesis.
- initiator tRNA
Special tRNA that intiates translation. It always carries the amino acid methionine.
- innate immune response
Immune response (of both vertebrates and invertebrates) to a pathogen that involves the pre-existing defenses of the body—the innate immune system—such as barriers formed by skin and mucosa, antimicrobial molecules and phagocytes. Such a response is not specific for the pathogen.
- inner membrane
The innermost of two membranes surrounding an organelle. In the mitochondrion, it encloses the matrix and contains the respiratory electron transport chains.
- inner nuclear membrane
The innermost of the two nuclear membranes. It contains binding sites for chromatin and the nuclear lamina on its internal face.
- inositol phospholipids (phosphoinositides)
One of a family of lipids containing phosphorylated inositol derivatives. Although minor components of the plasma membrane, they are important in signal transduction in eucaryotic cells. (See Figure 15–34.)
- insulator element
DNA sequence that prevents a gene regulatory protein bound to DNA in the control region of one gene from influencing the transcription of adjacent genes.
- insulin
Polypeptide hormone that is secreted by β cells in the pancreas and helps regulate glucose metabolism in animals.
- integral membrane protein
Protein that is held tightly in a membrane and can only be removed by treatments that disrupt the lipid bilayer.
- integrin
Member of a large family of transmembrane proteins involved in the adhesion of cells to the extracellular matrix and to each other.
- intercalary regeneration
Type of regeneration that fills in the missing tissues when two mismatched parts of a structure are grafted together.
- interferon-γ (IFN-)
Cytokine secreted by certain types of T cells after activation, and which enhances the anti-viral response and macrophage activation.
- interleukin
Secreted peptide or protein that mainly mediates local interactions between white blood cells (leucocytes) during inflammation and immune responses.
- intermediate filament
Fibrous protein filament (about 10 nm in diameter) that forms ropelike networks in animal cells. One of the three most prominent types of cytoskeletal filaments. (See Panel 16–1, p. 909.)
- intermembrane space
The subcompartment formed between the inner and outer mitochondrial membranes.
The corresponding compartment in a chloroplast.
- internal membrane
Eucaryotic cell membrane other than the plasma membrane. The membranes of the endoplasmic reticulum and the Golgi apparatus are examples.
- interphase
Long period of the cell cycle between one mitosis and the next. Includes G1 phase, S phase, and G2 phase.
- intracellular signaling protein
Protein that relays a signal as part of an intracellular signaling pathway. It may either activate the next protein in the pathway or generate a small intracellular mediator.
- intron
Noncoding region of a eucaryotic gene that is transcribed into an RNA molecule but is then excised by RNA splicing during production of the messenger RNA or other functional structural RNA.
- inversion
Type of mutation in which a segment of chromosome is inverted.
- ion channel
Transmembrane protein complex that forms a water-filled channel across the lipid bilayer through which specific inorganic ions can diffuse down their electrochemical gradients.
- ion
An atom that has either gained or lost electrons to acquire a charge; for example Na+ and Cl–.
- ionic bond
Cohesion between two atoms, one with a positive charge, the other with a negative charge. One type of noncovalent bond.
- ionophore
Small hydrophobic molecule that dissolves in lipid bilayers and increases their permeability to specific inorganic ions.
- iron-sulfur center
Electron-transporting group consisting of either two or four iron atoms bound to an equal number of sulfur atoms, found in a class of electron-transport proteins.
- isoelectric point
The pH at which a charged molecule in solution has no net electric charge and therefore does not move in an electric field.
- isomers
Molecules that are formed from the same atoms in the same chemical linkages but have different three-dimensional conformations. (See Panel 2–4, pp. 116–117.)
- isoprenoid (polyisoprenoid)
Member of a large family of lipid molecules with a carbon skeleton based on multiple five-carbon isoprene units. Examples include retinoic acid and dolichol.
- isotope
One of a number of forms of an atom that differ in atomic weight but have the same number of protons and electrons, and therefore the same chemistry. May be either stable or radioactive.
- Jak-STAT signaling pathway
Rapid signaling pathway by which some extracellular signals (for example interferon) activate gene expression. Involves cell-surface receptors and cytoplasmic Janus kinases (Jaks) plus signal transducers and activators of transcription (STATs).
- joule
Standard unit of energy in the meter-kilogram system. One joule is the energy delivered in one second by a one-watt power source. Approximately equal to 0.24 calories.
- K
see equilibrium constant
- K+ leak channel
A K+-transporting ion channel in the plasma membrane of animals cells that remains open even in a “resting” cell.
- karyotype
Full set of chromosomes of a cell arranged with respect to size, shape, and number.
- Ka
see affinity constant
- keratin
Member of the family of proteins that form keratin intermediate filaments, mainly in epithelial cells. Specialized keratins are found in hair, nails, and feathers.
- ketone
Organic molecule containing a carbonyl group linked to two alkyl groups.
- kilo-Prefix denoting 103
.
- kilocalorie (kcal)
Unit of heat energy equal to 1000 calories. Often used to express the energy content of food or molecules: bond strengths, for example, are measured in kcal/mole. An alternative unit in wide use is the kilojoule, equal to 0.24 kcal.
- kilojoule
Standard unit of energy equal to 1000 joules, or 0.24 kilocalories.
- kinesin
One type of motor protein that uses the energy of ATP hydrolysis to move along a microtubule.
- kinetochore
Complex structure formed from proteins on a mitotic chromosome to which microtubules attach and which plays an active part in the movement of chromosomes to the poles. The kinetochore forms on the part of the chromosome known as the centromere.
- kinetochore microtubule
In a mitotic or meiotic spindle, a microtubule with one end attached to the kinetochore on a chromosome.
- Krebs cycle
see citric acid cycle
- label
Chemical group, radioactive atom or fluorescent dye added to a molecule in order to follow its progress through a biochemical reaction or to locate it spatially. Also, as a verb, to add such a group or atom to a cell or molecule.
- lagging strand
One of the two newly synthesized strands of DNA found at a replication fork. The lagging strand is made in discontinuous lengths that are later joined covalently.
- lambda bacteriophage ( bacteriophage)
Virus that infects E. coli. Widely used as a DNA cloning vector
- lamellipodium (lamellipodia)
Flattened, sheetlike protrusion supported by a meshwork of actin filaments, which is extended at the leading edge of a crawling animal cell.
- laminin
Extracellular matrix protein found in basal laminae, where it forms a sheetlike network.
- lamin
see nuclear lamin
- lampbrush chromosome
Paired chromosome in meiosis in immature amphibian eggs, in which the chromatin forms large stiff loops extending out from the linear axis of the chromosome.
- leading strand
One of the two newly synthesized strands of DNA found at a replication fork. The leading strand is made by continuous synthesis in the 5′-to-3′ direction.
- lectin
Protein that binds tightly to a specific sugar. Abundant lectins from plant seeds are often used as affinity reagents to purify glycoproteins or to detect them on the surface of cells.
- leptotene
The first phase of division I of meiosis, in which the paired duplicated homologous chromosomes condense and become visible in the light microscope.
- lethal mutation
A mutation that causes the death of the cell or the organism that contains it.
- leucine zipper
Structural motif seen in many DNA-binding proteins in which two α helices from separate proteins are joined together in a coiled-coil (rather like a zipper), forming a protein dimer.
- leucine-rich repeat protein (LRR protein)
Common type of receptor serine/threonine kinase in plants. Characterized by a tandem array of leucine-rich repeat sequences in the extracellular portion.
- leucocyte
see white blood cell
- leukemia
Cancer of white blood cells.
- ligand
Any molecule that binds to a specific site on a protein or other molecule. (From Latin ligare, to bind.)
- ligase
Enzyme that joins together (ligates) two molecules in an energy-dependent process. DNA ligase, for example, joins two DNA molecules together end to end through phosphodiester bonds.
- light chain
One of the smaller polypeptides of a multisubunit protein such as myosin or immunoglobulin. Abbreviated as L chain in immunoglobulins.
- lineage analysis
Tracing the ancestry of individual cells in a developing embryo.
- linkage
Mutual effect of the binding of one ligand on the binding of another that is a central feature of the behavior of all allosteric proteins.
Co-inheritance of two genetic loci that lie near each other on the same chromosome. The closer together the two loci, that is, the greater the linkage, the lower the frequency of recombination between them.
- lipase
Enzyme that catalyzes the cleavage of fatty acids from the glycerol moiety of a triglyceride.
- lipid
Organic molecule that is insoluble in water but tends to dissolve in nonpolar organic solvents. A special class, the phospholipids, forms the structural basis of biological membranes.
- lipid bilayer
Thin bimolecular sheet of mainly phospholipid molecules that forms the core structure of all cell membranes. The two layers of lipid molecules are packed with their hydrophobic tails pointing inward and their hydrophilic heads outward, exposed to water.
- lipid raft
Small region of the plasma membrane enriched in sphingolipids and cholesterol.
- lipophilic
see hydrophobic
- liposome
Artificial phospholipid bilayer vesicle formed from an aqueous suspension of phospholipid molecules.
- local mediator
Secreted signal molecule that acts at short range on adjacent cells.
- locus
In genetics, the position of a gene on a chromosome. Different alleles of the same gene all occupy the same locus.
- long-term potentiation
Long-lasting increase (days to weeks) in the sensitivity of certain synapses in the hippocampus. Induced by a short burst of repetitive firing in the presynaptic neurons.
- low-density lipoprotein (LDL)
Large complex composed of a single protein molecule and many esterified cholesterol molecules, together with other lipids. The form in which cholesterol is transported in the blood and taken up into cells.
- LTP
- lumen
Cavity enclosed by an epithelial sheet (in a tissue) or by a membrane (in a cell).
- lymph
Colorless fluid derived from blood by filtration through capillary walls. Carries lymphocytes in a special system of ducts and vessels—the lymphatic vessels.
- lymphocyte
Type of white blood cell responsible for the specificity of adaptive immune responses. There are two main types: B cells, which produce antibody, and T cells, which interact directly with other effector cells of the immune system and with infected cells. T cells develop in the thymus and are responsible for cell-mediated immunity. B cells develop in the bone marrow in mammals and are responsible for the production of circulating antibodies.
- lymphoid organ
Organs involved in the production or function of lymphocytes, such as thymus, spleen, lymph nodes, and tonsils.
- lysis
Rupture of a cell’s plasma membrane, leading to the release of cytoplasm and the death of the cell.
- lysogeny
State of a bacterium in which it carries the DNA of an inactive virus integrated into its genome. The virus can subsequently be activated to replicate and lyse the cell.
- lysosome
Membrane-bounded organelle in eucaryotic cells containing digestive enzymes, which are typically most active at the acid pH found in the lumen of lysosomes.
- lysozyme
Enzyme that catalyzes the cutting of polysaccharide chains in the cell walls of bacteria.
- M phase
Period of the eucaryotic cell cycle during which the nucleus and cytoplasm divide.
- M6P
see mannose 6-phosphate
- macromolecule
Molecule such as a protein, nucleic acid, or polysaccharide with a molecular mass greater than a few thousand daltons.
- macrophage
Phagocytic cell derived from blood monocytes, typically resident in most tissues. It has both scavenger and antigen-presenting functions in immune responses.
- major histocompatibility complex (MHC)
Complex of highly polymorphic genes in vertebrates. They code for a large family of cell-surface glycoproteins (MHC molecules) that bind peptide fragments of foreign proteins and present them to T cells to induce an immune response. (See Figure 24–50.)
- malaria
Potentially fatal human disease caused by the protozoan parasite Plasmodium, which is transmitted by the bite of an infected mosquito.
- malignant
Describes tumors and tumor cells that are invasive and/or able to undergo metasis. A malignant tumor is a cancer.
- mannose 6-phosphate (M6P)
Unique marker attached to the oligosaccharides on some glycoproteins destined for lysosomes.
- map unit
see genetic map distance
- MAP
- MAP-kinase (mitogen-activated protein kinase)
Protein kinase that performs a crucial step in relaying signals from the plasma membrane to the nucleus. Turned on by a wide range of proliferation- or differentiation-inducing signals.
- mating-type locus (MAT locus)
In budding yeast, the locus that determines the mating type (α or a) of the haploid yeast cell.
- matrix space
Central subcompartment of a mitochondrion, bounded by the inner mitochondrial membrane.
The corresponding compartment in a chloroplast, which is more commonly known as the stroma.
- M-Cdk
see M-phase Cdk
- Mcm proteins
Proteins in the eucaryotic cell that bind to origin recognition complexes in DNA in early G1 and are involved in forming the pre-replicative complex.
- M-cyclin
Type of cyclin found in all eucaryotic cells that promotes the events of mitosis.
- MDR protein
see multidrug resistance protein
- mega-
Prefix denoting 106. (From Greek megas, huge, powerful.)
- megakaryocyte
Large myeloid cell with a multilobed nucleus that remains in the bone marrow when mature. It buds off platelets from long cytoplasmic processes.
- meiosis
Special type of cell division by which eggs and sperm cells are produced. It comprises two successive nuclear divisions with only one round of DNA replication, which produces four haploid daughter cells from an initial diploid cell.
- melanocyte
Cell that produces the dark pigment melanin. Responsible for the pigmentation of skin and hair.
- membrane
The lipid bilayer plus associated proteins that encloses all cells and, in eucaryotic cells, many organelles as well.
- membrane-bound ribosome
Ribosome attached to the cytosolic face of the endoplasmic reticulum. The site of synthesis of proteins that enter the endoplasmic reticulum.
- membrane channel
Transmembrane protein complex that allows inorganic ions or other small molecules to diffuse passively across the lipid bilayer.
- membrane potential
Voltage difference across a membrane due to a slight excess of positive ions on one side and of negative ions on the other. A typical membrane potential for an animal cell plasma membrane is –60 mV (inside negative relative to the surrounding fluid).
- membrane protein
Protein that is normally closely associated with a cell membrane. (See Figure 10–17.)
- membrane transport
Movement of molecules across a membrane mediated by a membrane transport protein.
- membrane transport protein
Membrane protein that mediates the passage of ions or molecules across a membrane. Examples are ion channels and carrier proteins.
- meristem
An organized group of dividing cells whose derivatives give rise to the tissues and organs of a flowering plant. Key examples are the apical meristems at the tips of shoots and roots.
- mesenchyme
Immature, unspecialized form of connective tissue in animals, consisting of cells embedded in a thin extracellular matrix.
- mesoderm
Embryonic tissue that is the precursor to muscle, connective tissue, skeleton and many of the internal organs.
- messenger RNA (mRNA)
RNA molecule that specifies the amino acid sequence of a protein. Produced by RNA splicing (in eucaryotes) from a larger RNA molecule made by RNA polymerase as a complementary copy of DNA. It is translated into protein in a process catalyzed by ribosomes.
- metabolism
The sum total of the chemical processes that take place in living cells.
- metaphase
Stage of mitosis at which chromosomes are firmly attached to the mitotic spindle at its equator but have not yet segregated toward opposite poles.
- metaphase plate
Imaginary plane at right angles to the mitotic spindle and midway between the spindle poles; the plane in which chromosomes are positioned at metaphase.
- metaplasia
A change in the pattern of cell differentiation in a tissue.
- metastasis
Spread of cancer cells from their site of origin to other sites in the body.
- methyl (CH3)
Hydrophobic chemical group derived from methane (CH4).
- MHC molecule
One of a large family of ubiquitous cell-surface glycoproteins encoded by genes of the major histocompatibility complex (MHC). They bind peptide fragments of foreign antigens and present them to T cells to induce an immune response. (See also class I MHC molecule, class II MHC molecule.)
- MHC
- micro-
Prefix denoting 10–6.
- microelectrode, micropipette
Piece of fine glass tubing pulled to an even finer tip. Used to penetrate a cell to study its physiology or to inject electric current or molecules.
- microfilament
see actin filament
- micrograph
Photograph of an image seen through a microscope. May be either a light micrograph or an electron micrograph depending on the type of microscope employed.
- microinjection
Injection of molecules into a cell using a micropipette.
- micron (m or micrometer)
Unit of measurement often applied to cells and organelles. Equal to 10–6 meter or 10–4 centimeter.
- micropipette
see microelectrode
- microsome
Small vesicle that is derived from fragmented endoplasmic reticulum produced when cells are homogenized.
- microtubule
Long hollow cylindrical structure composed of the protein tubulin. It is one of the three major classes of filaments of the cytoskeleton. (See Panel 16–1, p. 909.)
- microtubule-associated protein (MAP)
Any protein that binds to microtubules and modifies their properties. Many different kinds have been found, including structural proteins, such as MAP-2, and motor proteins, such as dynein.
- microtubule-organizing center (MTOC)
Region in a cell, such as a centrosome or a basal body, from which microtubules grow.
- microvillus (microvilli)
Thin cylindrical membrane-covered projection on the surface of an animal cell containing a core bundle of actin filaments. Present in especially large numbers on the absorptive surface of intestinal epithelial cells.
- midbody
Structure formed at the end of cleavage that can persist for some time as a tether between the two daughter cells in animals.
- milli-
Prefix denoting 10–3.
- minus end
The end of a microtubule or actin filament at which the addition of monomers occurs least readily; the “slow-growing” end of the microtubule or actin filament. The minus end of an actin filament is also known as the pointed end. (See Panel 16–2, pp. 912–913.)
- mismatch repair
DNA repair process that corrects mismatched nucleotides inserted during DNA replication. A short stretch of newly synthesized DNA including the mismatched nucleotide is removed and replaced with the correct sequence with reference to the template strand.
- mitochondrial precursor protein
Mitochondrial protein encoded by a nuclear gene, synthesized in the cytosol, and subsequently transported into mitochondria.
- mitochondrion (mitochondria)
Membrane-bounded organelle, about the size of a bacterium, that carries out oxidative phosphorylation and produces most of the ATP in eucaryotic cells.
- mitogen
An extracellular substance, such as a growth factor, that stimulates cell proliferation.
- mitogen-activated protein kinase
see MAP-kinase
- mitosis
Division of the nucleus of a eucaryotic cell, involving condensation of the DNA into visible chromosomes, and separation of the duplicated chromosomes to form two identical sets. (From Greek mitos, a thread, referring to the threadlike appearance of the condensed chromosomes.)
- mitotic chromosome
Highly condensed duplicated chromosome with the two new chromosomes still held together at the centromere as sister chromatids.
- mitotic spindle
Array of microtubules and associated molecules that forms between the opposite poles of a eucaryotic cell during mitosis and serves to move the duplicated chromosomes apart.
- model organism
A species, such as Drosophila melanogaster or Escherichia coli, that has been studied intensively over a long period and thus serves as a “model” of the biology of a particular type of organism.
- module
In proteins or nucleic acids, a unit of structure or function that is found in a variety of different contexts in different molecules.
- molar
Describes a solution with a concentration of 1 mole of a substance dissolved in 1 liter of solution (abbreviated as 1 M).
- mole
X grams of a substance, where X is its relative molecular mass (molecular weight). A mole consists of 6 × 1023 molecules of the substance.
- molecular chaperone
see chaperone
- molecular weight
Numerically, the same as the relative molecular mass of a molecule expressed in daltons. For example, a protein of relative molecular mass 20,000 has a molecular weight of 20,000.
- molecule
Group of atoms joined together by covalent bonds.
- monoclonal antibody
Antibody secreted by a hybridoma clone. Because each such clone is derived from a single B cell, all of the antibody molecules produced are identical.
- monocyte
Type of white blood cell that leaves the bloodstream and matures into a macrophage in tissues.
- monomer
Small molecular building block that can serve as a subunit, being linked to others of the same type to form a larger molecule (a polymer).
- monosaccharide
Simple sugar with the general formula (CH2O)n, where n = 3 to 8.
- morphogen
Signal molecule that can impose a pattern on a field of cells by causing cells in different places to adopt different fates.
- mosaic
In developmental biology, an organism made of a mixture of cells with different genotypes.
- motif
Element of structure or pattern that recurs in many contexts. Specifically, a small structural domain that can be recognized in a variety of proteins.
- motor protein
Protein that uses energy derived from nucleoside triphosphate hydrolysis to propel itself along a protein filament or another polymeric molecule.
- M-phase Cdk (M-Cdk)
Complex formed in vertebrate cells by an M-cyclin and the corresponding cyclin-dependent kinase (Cdk).
- mRNA
see messenger RNA
- MTOC
- multidrug resistance protein (MDR protein)
Type of ABC transporter protein that can pump hydrophobic drugs (such as some anti-cancer drugs) out of the cytoplasm of eucaryotic cells.
- multipass transmembrane protein
Membrane protein in which the polypeptide chain crosses the lipid bilayer more than once.
- mutant
Organism in which a mutation has occurred that makes it different from wild-type or from the ‘normal’ extent of variation in the population.
- mutation rate
The rate at which observable changes occur in a DNA sequence.
- mutation
Heritable change in the nucleotide sequence of a chromosome.
- myelin sheath
Insulating layer of specialized cell membrane wrapped around vertebrate axons. Produced by oligodendrocytes in the central nervous system and by Schwann cells in the peripheral nervous system.
- myeloid cell
Any white blood cell other than lymphocytes.
- myoblast
Mononucleated, undifferentiated muscle precursor cell. A skeletal muscle cell is formed by the fusion of multiple myoblasts.
- myoepithelial cell
Type of unstriated muscle cell found in epithelia, e.g. in the iris of the eye and in glandular tissue.
- myofibril
Long, highly organized bundle of actin, myosin, and other proteins in the cytoplasm of muscle cells that contracts by a sliding filament mechanism.
- N terminus
see amino terminus
- Na+-K+ pump (Na+-K+ ATPase)
Transmembrane carrier protein found in the plasma membrane of most animal cells that pumps Na+ out of and K+ into the cell, using energy derived from ATP hydrolysis.
- NAD+ (nicotine adenine dinucleotide)
Activated carrier that participates in an oxidation reaction by accepting a hydride ion (H–) from a donor molecule. The NADH formed is an important carrier of electrons for oxidative phosphorylation.
- NADH dehydrogenase complex
First of the three electron-driven proton pumps in the mitochondrial respiratory chain. It accepts electrons from NADH.
- NADP+ (nicotine adenine dinucleotide phosphate)
Activated carrier closely related to NAD+ that is used extensively in biosynthetic, rather than catabolic, pathways. The reduced form is NADPH.
- nano-
Prefix denoting 10–9.
- nanometer (nm)
Unit of length commonly used to measure molecules and cell organelles. 1 nm = 10–3 micrometer (μm) = 10–9 meter.
- natural killer cell (NK cell)
Cytotoxic cell of the innate immune system that can kill virus-infected cells.
- N-CAM
see neural cell adhesion molecule
- negative control
Type of control of gene expression in which the active DNA-binding form of the regulatory protein turns the gene off.
- negative staining
Staining technique for use in the electron electron microscope in which a reverse, or negative, image of the object is created.
- Nernst equation
Quantitative expression that relates the equilibrium ratio of concentrations of an ion on either side of a permeable membrane to the voltage difference across the membrane. (See Panel 11–2, p. 634.)
- nerve cell
see neuron
- neural cell adhesion molecule (N-CAM)
Cell adhesion molecule of the immunoglobulin superfamily, expressed by many cell types including most nerve cells. It mediates Ca2+-independent cell-cell attachment in vertebrates.
- neural tube
Tube of ectoderm that will form the brain and spinal cord in a vertebrate embryo.
- neurite
Long process growing from a nerve cell in culture. A generic term that does not specify whether the process is an axon or a dendrite.
- neurofilament
Type of intermediate filament found in nerve cells.
- neuromuscular junction
Specialized chemical synapse between an axon terminal of a motor neuron and a skeletal muscle cell.
- neuron (nerve cell)
Cell with long processes specialized to receive, conduct, and transmit signals in the nervous system.
- neuropeptide
Peptide secreted by neurons as a signaling molecule either at synapses or elsewhere.
- neurotransmitter
Small signal molecule secreted by the presynaptic nerve cell at a chemical synapse to relay the signal to the postsynaptic cell. Examples include acetylcholine, glutamate, GABA, glycine, and many neuropeptides.
- neutron
Uncharged subatomic particle that forms part of an atomic nucleus.
- neutrophil
White blood cell that is specialized for the uptake of particulate material by phagocytosis and which enters tissues that become infected or inflamed.
- nicotine adenine dinucleotide phosphate
see NADP+
- nicotine adenine dinucleotide
see NAD+
- nitric oxide (NO)
Gaseous signal molecule in both animals and plants. In animals it regulates smooth muscle contraction, for example; in plants it is involved in responses to injury or infection.
- nitrogen cycle
The natural circulation of nitrogen between molecular nitrogen in the atmosphere, inorganic molecules in the soil, and organic molecules in living organisms.
- nitrogen fixation
Biochemical process carried out by certain bacteria that reduces atmospheric nitrogen (N2) to ammonia, leading eventually to various nitrogen-containing metabolites.
- nitrogenase complex
Complex of enzymes in nitrogen-fixing bacteria that catalyzes the reduction of atmospheric N2 to ammonia.
- NK cell
see natural killer cell
- N-linked oligosaccharide
Chain of sugars attached to a protein through the NH2 group of the side chain of an asparagine residue.
- NMR (nuclear magnetic resonance)
Resonant absorption of electromagnetic radiation at a specific frequency by atomic nuclei in a magnetic field, due to flipping of the orientation of their magnetic dipole moments. The NMR spectrum provides information about the chemical environment of the nuclei. Two-dimensional NMR is used widely to determine the three-dimensional structure of small proteins.
- nm
see nanometer
- noncovalent attraction
Chemical bond in which, in contrast to a covalent bond, no electrons are shared. Noncovalent bonds are relatively weak, but they can sum together to produce strong, highly specific interactions between molecules.
- noncyclic photophosphorylation
Photosynthetic process that produces both ATP and NADPH in plants and cyanobacteria.
- nonenveloped virus
Virus consisting of a nucleic acid core and protein capsid only.
- nonpolar (apolar)
Lacking any asymmetric accumulation of positive and negative charge. Nonpolar molecules are generally insoluble in water.
- nonsense-mediated mRNA decay
Mechanism for removing aberrant mRNAs containing in-frame internal stop codons before they can be translated.
- Northern blotting
Technique in which RNA fragments separated by electrophoresis are immobilized on a paper sheet. A specific RNA is then detected by hybridization with a labeled nucleic acid probe.
- NO
see nitric oxide
- Notch
Receptor protein involved in many instances of choice of cell fate in animal development, for example in the specification of nerve cells from ectodermal epithelium. Its ligands are cell-surface proteins such as Delta and Serrate.
- notochord
Stiff rod of mesoderm that runs along the back of all chordate embryos. In vertebrates it does not persist and becomes incorporated into the vertebral column.
- NSF
Protein with ATPase activity that disassembles a complex of a v-SNARE and a t-SNARE.
- nuclear envelope
Double membrane surrounding the nucleus. Consists of an outer and inner membrane and is perforated by nuclear pores.
- nuclear export signal
Sorting signal contained in the structure of molecules and complexes, such as RNA and new ribosomal subunits, that are transported from the nucleus to the cytosol through nuclear pore complexes.
- nuclear lamin
Protein subunit of the intermediate filaments of the nuclear lamina.
- nuclear lamina
Fibrous meshwork of proteins on the inner surface of the inner nuclear membrane. It is made up of a network of intermediate filaments formed from nuclear lamins.
- nuclear localization signal (NLS)
Signal sequences or signal patches found in proteins destined for the nucleus and which enable their selective transport into the nucleus from the cytosol through the nuclear pore complexes.
- nuclear magnetic resonance
see NMR
- nuclear pore complex
Large multiprotein structure forming a channel (the nuclear pore) through the nuclear envelope that allows selected molecules to move between nucleus and cytoplasm.
- nuclear receptor superfamily
Intracellular receptors for hydrophobic signal molecules such as steroids and retinoic acid. The receptor-ligand complex acts as a transcription factor in the nucleus.
- nuclear transport
Movement of macromolecules into or out of the nucleus mediated by nuclear transport receptors.
- nucleation
Critical stage in the assembly of a polymeric structure, such as a microtubule, at which a small cluster of monomers aggregates in the correct arrangement to initiate rapid polymerization. (See Panel 16–2, pp. 912–913.) More generally, the rate-limiting step in an assembly process.
- nucleic acid
RNA or DNA, a macromolecule consisting of a chain of nucleotides joined together by phosphodiester bonds.
- nucleolar organizer
Region of a chromosome containing a cluster of ribosomal RNA genes that gives rise to a nucleolus.
- nucleolus
Structure in the nucleus where ribosomal RNA is transcribed and ribosomal subunits are assembled.
- nucleoporin
Any of a number of different proteins that make up nuclear pore complexes.
- nucleoside
Molecule composed of a purine or pyrimidine base covalently linked to a ribose or deoxyribose sugar. (See Panel 2–6, pp. 120–121.)
- nucleosome
Beadlike structure in eucaryotic chromatin. It is composed of a short length of DNA wrapped around a core of histone proteins, and is the fundamental structural unit of chromatin.
- nucleotide
Nucleoside with one or more phosphate groups joined in ester linkages to the sugar moiety. DNA and RNA are polymers of nucleotides. (See Panel 2–6, pp. 120–121.)
- nucleus
Prominent membrane-bounded organelle in a eucaryotic cell, containing DNA organized into chromosomes.
- nurse cell
Cell connected by cytoplasmic bridges to a developing oocyte and which thereby supplies it with ribosomes, mRNAs, and proteins needed for the development of the early embryo.
- occluding junction
Type of cell junction that seals cells together in an epithelium, forming a barrier through which even small molecules cannot pass.
- Okazaki fragments
Short lengths of DNA produced on the lagging strand during DNA replication. They are rapidly joined by DNA ligase to form a continuous DNA strand.
- oligodendrocyte
Type of glial cell in the vertebrate central nervous system that forms a myelin sheath around axons.
- oligomer
Short polymer, usually consisting (in a cell) of amino acids (oligopeptides), sugars (oligosaccharides), or nucleotides (oligonucleotides). (From Greek oligos, few, little.)
- oligosaccharide
Short linear or branched chain of covalently linked sugars (see Panel 2–4, pp. 116–117.) (See also complex oligosaccharide, high-mannose oligosaccharide, N-linked oligosacharide, O-linked glycosylation.)
- O-linked glycosylation
Addition of an oligosaccharide chain to a protein through the OH group of a serine or threonine side chain.
- oncogene
An altered gene whose product can act in a dominant fashion to help make a cell cancerous. Typically, an oncogene is a mutant form of a normal gene (proto-oncogene) involved in the control of cell growth or division.
- oocyte
The developing egg. It is usually a large and immobile cell.
- oogenesis
Formation and maturation of oocytes in the ovary.
- operator
Short region of DNA in a bacterial chromosome that controls the transcription of an adjacent gene.
- operon
In a bacterial chromosome, a group of contiguous genes that are transcribed into a single mRNA molecule.
- ORC
- organelle
Membrane-enclosed compartment in a eucaryotic cell that has a distinct structure, macromolecular composition, and function. Examples are nucleus, mitochondrion, chloroplast, Golgi apparatus.
- Organizer
- origin recognition complex (ORC)
Large protein complex that is bound to the DNA at origins of replication in eucaryotic chromosomes throughout the cell cycle.
- osmolarity
A term used to describe the concentration of a solute in terms of the osmotic pressure it can exert.
- osmosis
Net movement of water molecules across a semipermeable membrane driven by a difference in concentration of solute on either side. The membrane must be permeable to water but not to the solute molecules.
- osteoblast
Cell that secretes matrix of bone.
- osteoclast
Macrophage-like cell that erodes bone, enabling it to be remodeled during growth and in response to stresses throughout life.
- outer membrane
Outermost of the two membranes surrounding an organelle; the membrane adjacent to the cytosol.
- outer nuclear membrane
The outermost of the two nuclear membranes. It is continuous with the endoplasmic reticulum and is studded with ribosomes on its cytosolic face.
- overlap microtubule
In the mitotic or meiotic spindle, a microtubule interdigitating at the equator with the microtubules emanating from the other pole.
- ovulation
Release of an egg from the ovary.
- ovum
see egg
- oxidation (verb oxidize)
Loss of electrons from an atom, as occurs during the addition of oxygen to a molecule or when a hydrogen is removed. Opposite of reduction. (See Figure 2–43.)
- oxidative phosphorylation
Process in bacteria and mitochondria in which ATP formation is driven by the transfer of electrons from food molecules to molecular oxygen. Involves the intermediate generation of a proton gradient (pH gradient) across a membrane and chemiosmotic coupling.
- p53
Tumor suppressor gene found mutated in about half of human cancers. It encodes a gene regulatory protein that is activated by damage to DNA and is involved in blocking further progression through the cell cycle.
- pachytene
Third stage of division I of meiosis, in which synapsis is complete.
- palindromic sequence
Nucleotide sequence that is identical to its complementary strand when each is read in the same chemical direction—for example, GATC.
- paracrine signaling
Short-range cell-cell communication via secreted signal molecules that act on adjacent cells.
- parthenogenesis
Production of a new individual from an egg cell in the absence of fertilization by sperm.
- passive transport
Transport of a solute across a membrane down its concentration gradient or its electrochemical gradient, using only the energy stored in the gradient.
- patch-clamp recording
Electrophysiological technique in which a tiny electrode tip is sealed onto a patch of cell membrane, thereby making it possible to record the flow of current through individual ion channels in the patch.
- pathogen
(adjective pathogenic) An organism or other agent that causes disease.
- PCR (polymerase chain reaction)
Technique for amplifying specific regions of DNA by the use of sequence-specific primers and multiple cycles of DNA synthesis, each cycle being followed by a brief heat treatment to separate complementary strands.
- peptide bond
Chemical bond between the carbonyl group of one amino acid and the amino group of a second amino acid—a special form of amide linkage. Peptide bonds link amino acids together in proteins. (See Panel 3–1, pp. 132–133.)
- peptide map
Characteristic two-dimensional pattern (on paper or gel) formed by the separation of the mixture of peptides produced by the partial digestion of a protein.
- peripheral lymphoid organ (secondary lymphoid organ)
Lymphoid organ in which T cells and B cells interact with foreign antigens. Examples are spleen, lymph nodes, and mucosal-associated lymphoid tissue.
- peripheral membrane protein
Protein that is attached to one face of a membrane by noncovalent interactions with other membrane proteins, and which can be removed by relatively gentle treatments that leave the lipid bilayer intact.
- peroxisome
Small membrane-bounded organelle that uses molecular oxygen to oxidize organic molecules. Contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
- pH
Common measure of the acidity of a solution: “p” refers to power of 10, “H” to hydrogen. Defined as the negative logarithm of the hydrogen ion concentration in moles per liter (M). Thus on the pH scale, pH 3 (10–3 M H+) is acidic and pH 9 (10–9 M H+) is alkaline.
- PH domain
see pleckstrin homology domain
- phage display
Technique for detecting proteins that interact with each other by screening a protein against a library of genetically modified phage, each displaying a potential binding protein on their surface.
- phage
see bacteriophage
- phagocyte
General term for a professional phagocytic cell—that is, a cell such as a macrophage or neutrophil that is specialized to take up particles and microorganisms by phagocytosis.
- phagocytosis
Process by which particulate material is endocytosed (“eaten”) by a cell. Prominent in carnivorous cells, such as Amoeba proteus, and in vertebrate macrophages and neutrophils. (From Greek phagein, to eat.)
- phagosome
Large intracellular membrane-bounded vesicle that is formed as a result of phagocytosis. Contains ingested extracellular material.
- phase-contrast microscope
Type of light microscope that exploits the interference effects that occur when light passes through material of different refractive indexes. Used to view living cells.
- phenotype
The observable character of a cell or an organism.
- phosphatase
Enzyme that removes phosphate groups from a molecule.
- phosphatidylinositol 3-kinase (PI 3-kinase)
A kinase involved in intracellular signaling pathways activated by a variety of cell-surface receptors. It phosphorylates inositol phospholipids at the 3 position of the inositol ring. (See Figure 15–58.)
- phosphatidylinositol
An inositol phospholipid. (See Figure 15–34.)
- phosphodiester linkage
Set of covalent chemical bonds formed when two hydroxyl groups are linked in ester linkage to the same phosphate group. This linkage joins adjacent nucleotides in RNA or DNA. (See Figure 2–28.)
- phosphoinositide
see inositol phospholipid
- phospholipase C-β (PLC-)
Enzyme bound to the cytoplasmic face of the plasma membrane that converts membrane phosphatidylinositol 4,5-bisphosphate to diacylglycerol (which remains in the plasma membrane) and inositol 1,4,5-trisphosphate (IP3). It is activated by certain G proteins to trigger the inositol phospholipid signaling pathway.
- phospholipase C-γ (PLC-)
Like phospholipase C-β, an enzyme that cleaves inositol phospholipids to diacylglycerol and IP3 to trigger the inositol phospholipid signaling pathway. Activated by certain receptor tyrosine kinases.
- phospholipid exchange protein
Water-soluble carrier protein that transfers a phospholipid molecule from one membrane to another.
- phospholipid
The main category of lipid molecules used to construct biological membranes. Generally composed of two fatty acids linked through glycerol phosphate to one of a variety of polar groups.
- phosphoprotein phosphatase
Enzyme that removes a phosphate group from a protein by hydrolysis.
- phosphorylation
Reaction in which a phosphate group becomes covalently coupled to another molecule.
- photochemical reaction center
The part of a photosystem that converts light energy into chemical energy.
- photon
Elementary particle of light and other electromagnetic radiation.
- photoreceptor
Cell or molecule that is sensitive to light.
- photosynthesis
Process by which plants, algae and some bacteria use the energy of sunlight to drive the synthesis of organic molecules from carbon dioxide and water.
- photosynthetic electron-transfer
Light-driven reactions in photosynthesis in which electrons move along the electron-transport chain in the thylakoid membrane, generating ATP and NADPH.
- photosystem
Multiprotein complex involved in photosynthesis that captures the energy of sunlight and converts it to useful forms of energy.
- phragmoplast
Structure made of microtubules and actin filaments that forms in the prospective plane of division of a plant cell and guides formation of the cell plate.
- phylogeny
Evolutionary history of an organism or group of organisms, often presented in chart form as a phylogenetic tree.
- pinocytosis
Type of endocytosis in which soluble materials are taken up from the environment and incorporated into vesicles for digestion. Literally, “cell drinking.” (See also fluid-phase endocytosis.)
- PKA
see cyclic AMP-dependent protein kinase
- PKC
see protein kinase C
- plant growth regulator
Signal molecule (also known as a plant hormone) that helps coordinate growth and development. Examples are ethylene, auxins, gibberellins, cytokines, abscisic acid, and the brassinosteroids.
- plasma membrane
Membrane that surrounds a living cell.
- plasmid
Small circular DNA molecule that replicates independently of the genome. Modified plasmids are used extensively as plasmid vectors for DNA cloning.
- plasmodesma (plasmodesmata)
Communicating cell–cell junction in plants in which a channel of cytoplasm lined by plasma membrane connects two adjacent cells through a small pore in their cell walls.
- plastid
Cytoplasmic organelle in plants, bounded by a double membrane, that carries its own DNA and is often pigmented. Chloroplasts are plastids.
- platelet
Cell fragment, lacking a nucleus, that breaks off from a megakaryocyte in the bone marrow and is found in large numbers in the bloodstream. It helps initiate blood clotting when blood vessels are injured.
- PLC-β
see phospholipase C-β
- PLC-γ
see phospholipase C-γ
- pleckstrin homology domain (PH domain)
Protein domain found in intracellular signaling proteins by which they bind to inositol phospholipids phosphorylated by PI 3-kinase.
- ploidy
The number of complete sets of chromosomes in a genome. Diploid organisms have two sets in their somatic cells, polyploid organisms more than two. Natural polyploidy is the result of previous duplications of the whole genome or the introduction of complete genomes from another species during evolution.
- plus end
The end of a microtubule or actin filament at which addition of monomers occurs most readily; the “fast-growing” end of a microtubule or actin filament. The plus end of an actin filament is also known as the barbed end. (See Panel 16–2, pp. 912–913.)
- point mutation
Change of a single nucleotide in DNA, especially in a region of DNA coding for protein.
- polar
In the electrical sense, describes a structure (for example, a chemical bond, chemical group, or molecule) with positive charge concentrated toward one end and negative charge toward the other as a result of an uneven distribution of electrons. Polar molecules are likely to be soluble in water.
- polyisoprenoid–
see isoprenoid
- polymer
Large molecule made by covalently linking multiple identical or similar units (monomers) together.
- polymerase chain reaction
see PCR
- polymorphic
Describes a gene with many different alleles, none of which is predominant in the population.
- polypeptide
Linear polymer composed of multiple amino acids. Proteins are large polypeptides, and the two terms can be used interchangeably.
- polypeptide backbone
The chain of repeating carbon and nitrogen atoms, linked by peptide bonds, in a polypeptide or protein. The side chains of the amino acids project from this backbone.
- polyploid
Describes a cell or an organism that contains more than two sets of homologous chromosomes.
- polyribosome (polysome)
Messenger RNA molecule to which are attached a number of ribosomes engaged in protein synthesis.
- polysaccharide
Linear or branched polymer of monosaccharides. They include glycogen, starch, hyaluronic acid, and cellulose.
- polytene chromosome
Giant chromosome in which the DNA has undergone repeated replication without separation into new chromosomes.
- position effect
Differences in gene expression that depend on the position of the gene on the chromosome and probably reflect differences in the state of the chromatin along the chromosome.
- positional information
Information supplied to or possessed by cells according to their position in a multicellular organism. A cell’s internal record of its positional information is called its positional value.
- positive control
Type of control of gene expression in which the active DNA-binding form of the regulatory protein turns the gene on.
- posterior
Situated toward the tail end of the body.
- posttranscriptional control
Any control on gene expression that is exerted at a stage after transcription has begun.
- posttranslational
Describes any process involving a protein that occurs after protein synthesis is completed.
- posttranslational modification
The enzyme-catalyzed change to a protein made after it is synthesized. Examples are acetylation, cleavage, glycosylation, methylation, phosphorylation, and prenylation.
- pre-B cell
Iimmediate precursor of a B cell.
- prenylation
Covalent attachment of an isoprenoid lipid group to a protein.
- preprophase band
Circumferential band of microtubules and actin filaments that forms around a plant cell under the plasma membrane prior to mitosis and cell division.
- primary immune response
Adaptive immune response to an antigen that is made on first encounter with that antigen.
- primary structure
Sequence of monomer units in a linear polymer, such as the amino acid sequence of a protein.
- primordial germ cell
Cell set aside early in embryonic development that is a precursor to germ cells that give rise to gametes.
- primosome
A complex of DNA primase and DNA helicase that is formed on the lagging strand during DNA replication, improving the efficiency of replication.
- prion
An infectious abnormal form of a normal protein that is replicated in the host by forcing the normal proteins of the same type to adopt the aberrant structure.
- prion disease
Transmissible spongiform encephalopathies such as Kreutzfeld–Jacob disease in humans, scrapie in sheep and bovine spongiform encephalopathy (BSE) in cattle, that are apparently caused and transmitted by abnormal forms of a protein (prions).
- probe
Defined fragment of RNA or DNA, radioactively or chemically labeled, used to locate specific nucleic acid sequences by hybridization.
- procaryote (prokaryote)
Single-celled microorganism whose cells lack a well-defined, membrane-enclosed nucleus. The procaryotes comprise two of the major domains of living organisms—the Bacteria and the Archaea.
- programmed cell death
see apoptosis
- prometaphase
Phase of mitosis preceding metaphase in which the nuclear envelope breaks down and chromosomes first attach to the spindle.
- promoter
Nucleotide sequence in DNA to which RNA polymerase binds to begin transcription.
- prophase
First stage of mitosis, during which the chromosomes are condensed but not yet attached to a mitotic spindle.
- protease (proteinase, proteolytic enzyme)
Enzyme such as trypsin that degrades proteins by hydrolyzing some of their peptide bonds.
- proteasome
Large protein complex in the cytosol with proteolytic activity that is responsible for degrading proteins that have been marked for destruction by ubiquitylation or by some other means.
- protein
The major macromolecular constituent of cells. A linear polymer of amino acids linked together by peptide bonds in a specific sequence.
- protein domain
Portion of a protein that has a tertiary structure of its own. Larger proteins are generally composed of several domains, each connected to the next by short flexible regions of polypeptide chain.
- protein glycosylation
Posttranslational addition of oligosaccharide side chains to a protein.
- protein kinase
Enzyme that transfers the terminal phosphate group of ATP to a specific amino acid of a target protein.
- protein kinase C (PKC)
Ca2+-dependent protein kinase that, when activated by diacylglycerol and an increase in the concentration of Ca2+, phosphorylates target proteins on specific serine and threonine residues.
- protein module
see module
- protein phosphatase
- protein phosphorylation
The covalent addition of a phosphate group to a side chain of a protein catalyzed by a protein kinase.
- protein translocator
Membrane-bound protein that mediates the transport of another protein across an organelle membrane.
- proteoglycan
Molecule consisting of one or more glycosaminoglycan (GAG) chains attached to a core protein.
- proteolysis
Degradation of a protein by hydrolysis at one or more of its peptide bonds.
- proteolytic enzyme
see protease
- protofilament
A linear chain of protein subunits joined end to end, which associates laterally with other protofilaments to form cytoskeletal components such as microtubules and intermediate filaments.
- proton
Positively charged subatomic particle that forms part of an atomic nucleus. Hydrogen has a nucleus composed of a single proton (H+).
- proton-motive force
Driving force that moves protons across a membrane as a result of an electrochemical proton gradient.
- proto-oncogene
Normal gene, usually concerned with the regulation of cell proliferation, that can be converted into a cancer-promoting oncogene by mutation.
- protozoa
Free-living or parasitic, nonphotosynthetic, single-celled, motile eucaryotic organisms, such as Paramecium and Amoeba. Free-living protozoa feed on bacteria or other microorganisms.
- pseudogene
Gene that has accumulated multiple mutations that has rendered it inactive and nonfunctional.
- pseudopodium (pseudopodia)
Large cell-surface protrusion formed by amoeboid cells as they crawl. More generally, any dynamic actin-rich extension of the surface of an animal cell.
- pulse-chase
Technique for following the movement of a substance through a biochemical or cellular pathway, by briefly adding the radioactively labeled substance (the pulse) followed by the unlabeled substance (the chase).
- pump
Transmembrane protein that drives the active transport of ions or small molecules across the lipid bilayer.
- purine
One of the two categories of nitrogen-containing ring compounds found in DNA and RNA. Examples are adenine and guanine. (See Panel 2–6, pp. 120–121.)
- pyrimidine
One of the two categories of nitrogen-containing ring compounds found in DNA and RNA. Cytosine, thymine and uracil are pyrimidines. (See Panel 2–6, pp. 120–121.)
- quaternary structure
Three-dimensional relationship of the different polypeptide chains in a multisubunit protein or protein complex.
- quinone (Q)
Small, lipid soluble, mobile electron carrier molecule found in the respiratory and photosynthetic electron-transport chains. (See Figure 14–24.)
- Rab protein
Any of a large family of monomeric GTPases present in the plasma membrane and organelle membranes that are involved in conferring specificity on vesicle docking.
- radioactive isotope
Form of an atom with an unstable nucleus that emits radiation as it decays.
- Ran
Monomeric GTPase present in both cytosol and nucleus that is required for the active transport of macromolecules into and out of the nucleus through nculear pore complexes. Hydrolysis of GTP to GDP is thought to provide the energy required for this transport.
- Ras protein
The most famous member of a large family of GTP-binding proteins (called monomeric GTPases) that help relay signals from cell-surface receptors to the nucleus. Named for the ras gene, first identified in viruses that cause rat sarcomas.
- reaction
In chemistry, any process in which one molecule is converted into another by the removal or addition of atoms, or in which the arrangement of atoms in a molecule or molecules is altered by a change in chemical bonds.
- reading frame
The phase in which nucleotides are read in sets of three to encode a protein. A messenger RNA molecule can be read in any one of three reading frames, only one of which will give the required protein. (See Figure 6–51.)
- RecA protein
The prototype for a class of DNA-binding proteins that catalyze synapsis of DNA strands during genetic recombination.
- receptor
Protein that binds a specific extracellular signal molecule (ligand) and initiates a response in the cell. Cell-surface receptors, such as the acetylcholine receptor and the insulin receptor, are located in the plasma membrane, with their ligand-binding site exposed to the external medium. Intracellular receptors, such as steroid hormone receptors, bind ligands that diffuse into the cell across the plasma membrane.
- receptor-mediated endocytosis
Internalization of receptor-ligand complexes from the plasma membrane by endocytosis, It is used to take up some macromolecules, such as cholesterol-containing lipoproteins, from the extracellular fluid, and is also a means of recycling receptor proteins once they have bound their ligands.
- recessive
In genetics, refers to the member of a pair of alleles that fails to be expressed in the phenotype of the organism when the dominant allele is present. Also refers to the phenotype of an individual that has only the recessive allele.
- recombinant DNA
Any DNA molecule formed by joining DNA segments from different sources. Recombinant DNAs are widely used in the cloning of genes, in the genetic modification of organisms, and in molecular biology generally.
- recombination
Process in which DNA molecules are broken and the fragments are rejoined in new combinations. Can occur in the living cell—for example, through crossing-over during meiosis—or in vitro using purified DNA and enzymes that break and ligate DNA strands.
- recycling endosomes
Large intracellular membrane-bounded vesicle formed from a fragment of an endosome that is an intermediate stage on the passage of recycled receptors back to the cell membrane.
- red blood cell
see erythrocyte
- redox pair
Pair of molecules in which one acts as an electron donor and one as an electron acceptor in an oxidation-reduction reaction; for example, NADH (electron donor) and NAD+ (electron acceptor).
- redox potential
The affinity of a redox pair for electrons, generally measured as the voltage difference between an equimolar mixture of the pair and a standard reference. NADH/NAD+ has a low redox potential and O2/H2 has a high redox potential (high affinity for electrons).
- redox reaction
A reaction in which one component becomes oxidized and the other reduced; an oxidation-reduction reaction.
- reduction (verb reduce)
Addition of electrons to an atom, as occurs during the addition of hydrogen to a molecule or the removal of oxygen from it. Opposite of oxidation. (See Figure 2–43.)
- regulatory sequence
DNA sequence to which a gene regulatory protein binds to control the rate of assembly of the transcirptional complex at the promoter.
- regulatory site
Site on an enzyme, other than the active site, that binds a molecule that affects enzyme activity.
- replication fork
Y-shaped region of a replicating DNA molecule at which the two daughter strands are formed and separate.
- replication origin
Location on a DNA molecule at which duplication of the DNA begins.
- replicative cell senescence
Phenomenon observed in primary cell cultures as they age, in which cell proliferation slows down and finally halts.
- repressor
Protein that binds to a specific region of DNA to prevent transcription of an adjacent gene.
- residue
General term for the unit of a polymer. That portion of a sugar, amino acid, or nucleotide that is retained as part of the polymer chain during the process of polymerization.
- respiration
General term for a process in a cell involving the oxidative breakdown of sugars or other organic molecules, and requiring the uptake of O2 while producing CO2 and H2O as waste products.
- respiratory chain
Electron-transport chain in the inner mitochondrial membrane that receives high-energy electrons derived from the citric acid cycle and generates the proton gradient across the membrane that is used to power ATP synthesis.
- respiratory control
Regulatory mechanism that controls the rate of electron transport in the respiratory chain according to need via a direct influence of the electrochemical proton gradient.
- respiratory enzyme complex
Any of the major protein complexes of the mitochondrial respiratory chain that act as electron-driven proton pumps to generate the proton gradient across the inner membrane.
- resting membrane potential
The membrane potential in equilibrium conditions in which there is no net flow of ions across the plasma membrane.
- restriction map
Diagrammatic representation of a DNA molecule indicating the sites of cleavage by various restriction enzymes.
- restriction nuclease (restriction enzyme)
One of a large number of nucleases that can cleave a DNA molecule at any site where a specific short sequence of nucleotides occurs. Extensively used in recombinant DNA technology.
- restriction point
Important checkpoint in the mammalian cell cycle. Passage through the restriction point commits the cell to enter S phase. It corresponds to Start in the yeast cell cycle.
- retrotransposon
Type of transposable element that moves by being first transcribed into an RNA copy that is then reconverted to DNA by reverse transcriptase and inserted elsewhere in the chromosomes.
- retrovirus
RNA-containing virus that replicates in a cell by first making a double-stranded DNA intermediate.
- reverse genetics
Approach to discovering gene function that starts from the DNA (gene) and protein and then creates mutants to analyze the gene’s function.
- reverse transcriptase
Enzyme first discovered in retroviruses that makes a double-stranded DNA copy from a single-stranded RNA template molecule.
- rhodopsin
G-protein-linked light-sensitive receptor protein in the rod photoreceptor cells of the retina.
- ribonuclease
Enzyme that cuts an RNA molecule by hydrolyzing one or more of its phosphodiester bonds.
- ribonucleic acid
see RNA
- ribosomal RNA (rRNA)
Any one of a number of specific RNA molecules that form part of the structure of a ribosome and participate in the synthesis of proteins. Often distinguished by their sedimentation coefficient, such as 28S rRNA or 5S rRNA.
- ribosome
Particle composed of ribosomal RNAs and ribosomal proteins that associates with messenger RNA and catalyzes the synthesis of protein.
- ribozyme
RNA with catalytic activity.
- RNA (ribonucleic acid)
Polymer formed from covalently linked ribonucleotide monomers. (See also messenger RNA, ribosomal RNA, transfer RNA.)
- RNA editing
Production of a functional mRNA by insertion or alteration of individual nucleotides in an RNA molecule after it is synthesized.
- RNA interference (RNAi)
Selective intracellular degradation of RNA that is intended to remove foreign RNAs, such as those of viruses. Fragments cleaved from free double-stranded RNA direct the degradative mechanism to other similar RNA sequences. Widely exploited in a technique used to silence the expression of selected genes.
- RNA polymerase II holoenzyme
Large pre-assumbled complex of RNA polymerase II, most of the general transcription factors required for its function, and the mediator protein complex.
- RNA polymerase
Enzyme that catalyzes the synthesis of an RNA molecule on a DNA template from nucleoside triphosphate precursors. (See Figure 6–8.)
- RNA primer
Short stretch of RNA synthesized on a DNA template. It is required by DNA polymerases to start their DNA synthesis.
- RNA processing control
Control of gene expression by controlling how the RNA transcript is spliced or otherwise processed.
- RNA splicing
Process in which intron sequences are excised from RNA transcripts in the nucleus during formation of messenger and other RNAs.
- RNAi
see RNA interference
- rod photoreceptor (rod)
Photoreceptor cell type in the retina that is responsible for noncolor vision in dim light.
- rough endoplasmic reticulum (rough ER)
Endoplasmic reticulum with ribosomes on its cytosolic surface. Involved in the synthesis of secreted and membrane-bound proteins.
- rRNA
see ribosomal RNA
- rRNA gene
Gene that specifies a ribosomal RNA (rRNA).
- S phase
Period of a eucaryotic cell cycle in which DNA is synthesized.
- Saccharomyces
Genus of yeasts that reproduce asexually by budding or sexually by conjugation. Economically important in brewing and baking, they are also widely used in genetic engineering and as simple model organisms in the study of eucaryotic cell biology.
- sarcoma
Cancer of connective tissue.
- sarcomere
Repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick (myosin) and thin (actin) filaments between two adjacent Z discs.
- sarcoplasmic reticulum
Network of internal membranes in the cytoplasm of a muscle cell that contains high concentrations of sequestered Ca2+ which is released into the cytosol during muscle excitation.
- satellite DNA
Regions of highly repetitive DNA from a eucaryotic chromosome, usually identifiable by its unusual nucleotide composition. Satellite DNA is not transcribed and has no known function.
- saturated
Describes a molecule containing carbon–carbon bonds that has only single covalent bonds.
- scaffold protein
Protein that organizes groups of interacting intracellular signaling proteins into signaling complexes.
- scanning electron microscope
Type of electron microscope that produces an image of the surface of an object.
- S-Cdk
Complex formed in vertebrate cells by an S-cyclin and the corresponding cyclin-dependent kinase (Cdk).
- Schwann cell
Glial cell responsible for forming myelin sheaths in the peripheral nervous system.
- SDS polyacrylamide gel electrophoresis (SDS-PAGE)
Type of electrophoresis in which the protein mixture to be separated is run through a gel containing the detergent sodium dodecyl sulfate (SDS) which unfolds the proteins and frees them from association with other molecules.
- second messenger
Small molecule that is formed in or released into the cytosol in response to an extracellular signal and helps to relay the signal to the interior of the cell. Examples include cAMP, IP3, and Ca2+.
- secondary immune response
Adaptive immune response to an antigen that is made on a second or subsequent encounter with a given antigen. It is more rapid in onset, stronger, and more specific than the primary immune response.
- secondary structure
Regular local folding pattern of a polymeric molecule. In proteins, α helices and β sheets.
- secretory vesicle
Membrane-bounded organelle in which molecules destined for secretion are stored prior to release. Sometimes called secretory granule because darkly staining contents make the organelle visible as a small solid object.
- section
A very thin slice of tissue, suitable for viewing under the microscope.
- selectin
Member of a family of cell-surface carbohydrate-binding proteins that mediate transient, Ca2+-dependent cell-cell adhesion in the bloodstream, for example between white blood cells and the endothelium of the blood vessel wall.
- selectivity filter
That part of an ion channel structure that determines which ions it can transport.
- septate junction
Main type of occluding cell junction in invertebrates; their structure is distinct from that of vertebrate tight junctions.
- serine protease
Type of protease that has a reactive serine in the active site.
- sex chromosome
Chromosome that may be present or absent, or present in a variable number of copies, according to the sex of the individual. In mammals, the X and Y chromosomes.
- sexual reproduction
Type of reproduction in which the genomes of two individuals are mixed in the formation of a new organism. Individuals produced by sexual reproduction differ from either of their parents and from each other.
- SH2 domain
Src homology region 2, a protein domain present in many signaling proteins; it binds a short amino acid sequence containing a phosphotyrosine.
- side chain
The part of an amino acid that differs between different amino acids, giving the amino acid its unique physical and chemical properties.
- signal molecule
Extracellular or intracellular molecule that cues the response of a cell to the behavior of other cells or objects in the environment.
- signal patch
Protein sorting signal that consists of a specific three-dimensional arrangement of atoms on the folded protein’s surface.
- signal peptidase
Enzyme that removes a terminal signal sequence from a protein once the sorting process is complete.
- signal-recognition particle (SRP)
Ribonucleoprotein particle that binds an ER signal sequence on a partially synthesized polypeptide chain and directs the polypeptide and its attached ribosome to the endoplasmic reticulum.
- signal sequence
Short continuous sequence of amino acids that determines the eventual location of a protein in the cell. An example is the N-terminal sequence of 20 or so amino acids that directs nascent secretory and transmembrane proteins to the endoplasmic reticulum.
- signal transduction
Relaying of a signal by conversion from one physical or chemical form to another. In cell biology, the process by which a cell converts an extracellular signal into a response.
- single-nucleotide polymorphism (SNP)
Variation between individuals at certain nucleotide positions in the genome.
- single-pass transmembrane protein
Membrane protein in which the polypeptide chain crosses the lipid bilayer only once.
- single-strand DNA-binding protein
Protein that binds to the single strands of the opened-up DNA double helix, preventing helical structures from reforming while the DNA is being replicated.
- sister chromatid
see chromatid
- site-directed mutagenesis
Technique by which a mutation can be made at a particular site in DNA.
- site-specific recombination
Type of recombination that does not require extensive similarity in the two DNA sequences undergoing recombination. Can occur between two different DNA molecules or within a single DNA molecule.
- small intracellular mediator
see second messenger
- small nuclear RNA (snRNA)
Small RNA molecules that are complexed with proteins to form the ribonucleoprotein particles involved in RNA splicing.
- smooth endoplasmic reticulum (smooth ER)
Region of the endoplasmic reticulum not associated with ribosomes. It is involved in lipid synthesis.
- smooth muscle cell
Type of long, spindle-shaped mononucleate muscle cell making up the muscular tissue found in the walls of arteries and of the intestine and other viscera, and in some other locations of the vertebrate body. Called “smooth” because it lacks the striated myofibrils of skeletal and cardiac muscle cells.
- SNAREs
Large family of transmembrane proteins present in organelle membranes and the vesicles derived from them. They are involved in guiding vesicles to their correct destinations. They exist in pairs—a v-SNARE in the vesicle membrane that binds specifically to a complementary t-SNARE in the target membrane.
- SNP
- snRNA
see small nuclear RNA
- solute
Any molecule that is dissolved in a liquid. The liquid is called a solvent.
- somatic cell
Any cell of a plant or animal other than a germ cell or germ-cell precursor. (From Greek soma, body.)
- somite
One of a series of paired blocks of mesoderm that form during early development and lie on either side of the notochord in a vertebrate embryo. They give rise to the vertebral column, muscles and associated connective tissue. Each somite produces the musculature of one vertebral segment, plus associated connective tissue.
- sorting signal
Amino acid sequence that directs the delivery of a protein to a specific location outside the cytosol.
- Southern blotting
Technique in which DNA fragments separated by electrophoresis are immobilized on a paper sheet. Specific fragments are then detected with a labeled nucleic acid probe. (Named after E.M. Southern, inventor of the technique.)
- spectrin
Abundant protein associated with the cytosolic side of the plasma membrane in red blood cells, forming a rigid network that supports the membrane.
- Spemann’s Organizer
Specialized tissue at the dorsal lip of the blastopore in an amphibian embryo; a source of signals that help to orchestrate formation of the embryonic body axis. (After H. Spemann and H. Mangold, co-discoverers.)
- sperm (spermatozoon,spermatozoa)
The mature male gamete in animals. It is motile and usually small compared with the egg.
- spermatogenesis
Development of sperm.
- spindle-attachment checkpoint
Checkpoint that operates during mitosis to ensure that all chromosomes are properly attached to the spindle before sister-chromatid separation starts.
- spliceosome
Large assembly of RNA and protein molecules that performs pre-mRNA splicing in eucaryotic cells.
- Src family
Family of cytoplasmic tyrosine kinases (pronounced “sark”) that associate with the cytoplasmic domains of some enzyme-linked receptors (for example, the T cell antigen receptor) that lack intrinsic tyrosine kinase activity. They transmit a signal onwards by phosphorylating the receptor itself and other signaling proteins.
- SRP
- standard free-energy change (G)
Free-energy change of two reacting molecules at standard temperature and pressure when all components are present at a concentration of 1 mole per liter.
- starch
Polysaccharide composed exclusively of glucose units, used as an energy storage material in plant cells.
- start-transfer signal
Short amino-acid sequence that enables a polypeptide chain to start being translocated across the endoplasmic reticulum membrane through a protein translocator. Multipass membrane proteins have both N-terminal (signal sequence) and internal start-transfer signals.
- stem cell
Relatively undifferentiated cell that can continue dividing indefinitely, throwing off daughter cells that can undergo terminal differentiation into particular cell types.
- stereocilium
A large, rigid microvillus found in “organ pipe” arrays on the apical surface of hair cells in the ear. A stereocilium contains a bundle of actin filaments, rather than microtubules, and is thus not a true cilium.
- steroid
Hydrophobic lipid molecule with a characteristic four-ringed structure. Many important hormones such as estrogen and testosterone are steroids. (See Panel 2–5, pp. 118–119.)
- stimulatory G protein (Gs)
G protein that, when activated, activates the enzyme adenylyl cyclase and thus stimulates the production of cyclic AMP.
- stop-transfer signal
Hydrophobic amino acid sequence that halts translocation of a polypeptide chain through the endoplasmic reticulum membrane, thus anchoring the protein chain in the membrane (See Figure 12–49).
- strand-directed mismatch repair
see mismatch repair
- striated muscle
Muscle composed of transversely striped (striated) myofibrils. Skeletal and heart muscle of vertebrates are the best-known examples.
- stroma
The connective tissue in which a glandular or other epithelium is embedded.
The large interior space of a chloroplast, containing enzymes that incorporate CO2 into sugars.
- structural gene
Region of DNA that codes for a protein or for an RNA molecule that forms part of a structure or has an enzymatic function. Distinguished from regions of DNA that regulate gene expression.
- substrate
Molecule on which an enzyme acts.
- substratum
Solid surface to which a cell adheres.
- subunit
Component of a multicomponent complex—for example, one protein component of a protein complex or one polypeptide chain of a multichain protein.
- sucrose
Disaccharide composed of one glucose unit and one fructose unit. The major form in which glucose is transported between plant cells.
- sugar
Small carbohydrates with a monomer unit of general formula (CH2O)n. Examples are the monosaccharides glucose, fructose and mannose, and the disacharide sucrose (composed of a molecule of glucose and one of fructose linked together).
- sulfhydryl (thiol, SH)
Chemical group containing sulfur and hydrogen found in the amino acid cysteine and other molecules. Two sulfhydryls can join to produce a disulfide bond.
- supercoiled DNA
Region of DNA in which the double helix is further twisted on itself. (See Figure 6–20.)
- survival factor
Extracellular signal required for a cell to survive; in its absence the cell will undergo apoptosis and die.
- symbiosis
Intimate association between two organisms of different species from which both derive a long-term selective advantage.
- symporter
Carrier protein that transports two types of solute across the membrane in the same direction.
- synapse
Communicating cell–cell junction that allows signals to pass from a nerve cell to another cell. In a chemical synapse the signal is carried by a diffusible neurotransmitter; in an electrical synapse a direct connection is made between the cytoplasms of the two cells via gap junctions.
- synapsis
In genetic recombination, the initial formation of base pairs between complementary DNA strands in different DNA molecules that occurs at sites of crossing-over between chromosomes.
In meiosis, the pairing of maternal and paternal copies of a chromosome as they become attached to each other along their length.
- synaptic signaling
Type of cell–cell communication that occurs across chemical synapses in the nervous system.
- synaptic vesicle
Small neurotransmitter-filled secretory vesicle formed at the axon terminals of nerve cells and whose contents are released into the synaptic cleft by exocytosis when an action potential reaches the axon terminal.
- synaptonemal complex
Structure that holds paired chromosomes together during prophase I of meiosis and promotes genetic recombination.
- syncytium
Mass of cytoplasm containing many nuclei enclosed by a single plasma membrane. Typically the result either of cell fusion or of a series of incomplete division cycles in which the nuclei divide but the cell does not.
- synteny
The presence in different species of regions of chromosomes with the same genes in the same order.
- T cell (T lymphocyte)
Type of lymphocyte responsible for cell-mediated immunity; includes both cytotoxic T cells and helper T cells.
- TATA box
Consensus sequence in the promoter region of many eucaryotic genes that binds a general transcription factor and hence specifies the position at which transcription is initiated.
- TCA cycle
see citric acid cycle
- telomerase
- telomere
End of a chromosome, associated with a characteristic DNA sequence that is replicated in a special way. Counteracts the tendency of the chromosome otherwise to shorten with each round of replication. (From Greek telos, end.)
- telophase
Final stage of mitosis in which the two sets of separated chromosomes decondense and become enclosed by nuclear envelopes.
- temperature-sensitive (ts) mutant
Organism or cell carrying a genetically altered protein (or RNA molecule) that performs normally at one temperature but is abnormal at another (usually higher) temperature.
- template
A single strand of DNA or RNA whose nucleotide sequence acts as a guide for the synthesis of a complementary strand.
- terminator
Signal in bacterial DNA that halts transcription.
- tertiary structure
Complex three-dimensional form of a folded polymer chain, especially a protein or RNA molecule.
- TGF-β superfamily
see transforming growth factor-β superfamily
- TGN
see trans Golgi network (TGN)
- thioester bond
High-energy bond formed by a condensation reaction between an acid (acyl) group and a thiol group (–SH); seen, for example, in acetyl CoA and in many enzyme-substrate complexes.
- thiol
see sulfhydryl
- thylakoid
Flattened sac of membrane in a chloroplast that contains chlorophyll and other pigments and carries out the light-trapping reactions of photosynthesis. Stacks of thylakoids form the grana of chloroplasts.
- tight junction
Cell–cell junction that seals adjacent epithelial cells together, preventing the passage of most dissolved molecules from one side of the epithelial sheet to the other.
- TIM complexes
Protein translocators in the mitochondrial inner membrane. The TIM23 complex mediates the transport of proteins into the matrix and the insertion of some proteins into the inner membrane; the TIM22 complex mediates the insertion of a subgroup of proteins into the inner membrane.
- Toll-like receptor family (TLR)
Important family of mammalian pattern recognition receptors abundant on macrophages, neutrophils and the epithelial cells of the gut. They recognize pathogen-associated immunostimulants such as lipopolysacharide and peptidoglycan.
- TOM complex
Multisubunit protein complex that transports proteins across the mitochondrial outer membrane.
- topoisomerase (DNA topoisomerase)
Enzyme that makes reversible cuts in a double-helical DNA molecule for the purpose of removing knots or unwinding excessive twists.
- tracer
Molecule or atom that has been labeled either chemically or radioactively so that it can be followed in a biochemical process or readily located in a cell or tissue.
- trans face
Face of a Golgi stack at which material leaves the organelle for the cell surface or another cell compartment. It is adjacent to the trans Golgi network.
- trans Golgi network (TGN)
Network of interconnected cisternae and tubules at the trans face of the Golgi apparatus, through which material is transferred out of the Golgi.
- transcellular transport
Transport of solutes, such as nutrients, across an epithelium, by means of membrane transport proteins in the apical and basal faces of the epithelial cells.
- transcript
RNA product of DNA transcription.
- transcription (DNA transcription)
Copying of one strand of DNA into a complementary RNA sequence by the enzyme RNA polymerase.
- transcription attenuation
Inhibition of gene expression in bacteria by the premature termination of transcription.
- transcription factor
Term loosely applied to any protein required to initiate or regulate transcription in eucaryotes. Includes both gene regulatory proteins as well as the general transcription factors.
- transcriptional control
Control of of gene expression by controlling when and how often the gene is transcribed.
- transcytosis
The uptake of material at one face of a cell by endocytosis, its transfer across a cell in vesicles, and its discharge from another face by exocytosis.
- transfection
Introduction of a foreign DNA molecule into a eucaryotic cell. It is usually followed by expression of one or more genes in the newly introduced DNA.
- transfer RNA (tRNA)
Set of small RNA molecules used in protein synthesis as an interface (adaptor) between messenger RNA and amino acids. Each type of tRNA molecule is covalently linked to a particular amino acid.
- transforming growth factor-β superfamily (TGF- superfamily)
Large family of structurally related, secreted proteins that act as hormones and local mediators to control a wide range of functions in animals, including during development. It includes TGF-βs, activins, and bone morphogenetic proteins (BMPs).
- transgenic organism
Plant or animal that has stably incorporated one or more genes from another cell or organism and can pass them on to successive generations.
- transition state
Structure that forms transiently in the course of a chemical reaction and has the highest free energy of any reaction intermediate. Its formation is a rate-limiting step in the reaction.
- translation (RNA translation)
Process by which the sequence of nucleotides in a messenger RNA molecule directs the incorporation of amino acids into protein. It occurs on a ribosome.
- translational control
Control of gene expression by selection of which mRNAs in the cytoplasm are translated by ribosomes.
- translocation
Type of mutation in which a portion of one chromosome is broken off and attached to another.
- transmembrane protein
Membrane protein that extends through the lipid bilayer, with part of its mass on either side of the membrane.
- transmitter-gated ion channel
Ion channel in the postsynaptic plasma membranes of nerve and muscle cells that opens only in response to the binding of a specific extracellular neurotransmitter. The resulting inflow of ions leads to the generation of a local electrical signal in the postsynaptic cell.
- transposable element
Segment of DNA that can move from one position in a genome to another. Also called a transposon.
- transposition
The movement of a DNA sequence from one site to another within the genome. See also cut-and-paste transposition.
- trans-splicing
Type of RNA splicing present in a few eucaryotic organisms in which exons from two separate RNA molecules are joined together to form an mRNA.
- treadmilling
The process by which a polymeric protein filament is maintained at constant length by addition of protein subunits at one end and loss of subunits at the other. (See Panel 16–2, pp. 912–913.)
- triacylglycerol
Molecule composed of three fatty acids esterified to glycerol. The main constituent of fat droplets in animal tissues (where the fatty acids are saturated) and of vegetable oils (where the fatty acids are mainly unsaturated). Also known as triglyceride. (See Panel 2–5, pp. 118–119.)
- tricarboxylic acid (TCA) cycle
see citric acid cycle
- trimeric GTP-binding protein
see GTP-binding protein
- tRNA
see transfer RNA
- t-SNARE
see SNAREs
- tubulin
- tumor progression
The process by which an initial mildly disordered cell behavior gradually evolves into a full-blown cancer.
- tumor suppressor gene
Gene that appears to prevent formation of a cancer. Loss-of-function mutations in such genes enhance susceptibility to cancer.
- two-dimensional gel electrophoresis
Type of electrophoresis in which the protein mixture is run first in one direction and then in a direction at right angles to the first. It enables better separation of individual proteins.
- two-hybrid system
Technique for identifying interacting proteins using genetically engineered yeast cells.
- type III secretion system
A bacterial system for delivering toxic proteins into the cells of their host.
- ubiquitin
Small, highly conserved protein present in all eucaryotic cells that becomes covalently attached to lysines of other proteins. Attachment of a short chain of ubiquitins to such a lysine tags a protein for intracellular proteolytic destruction by a proteasome.
- ubiquitin ligase
Any one of a large number of enzymes that attach ubiquitin to a protein, thus marking it for destruction in a proteasome. The process catalyzed by a ubiquitin ligase is called ubiquitylation.
- unfolded protein response
Cellular response triggered by an accumulation of misfolded proteins in the endoplasmic reticulum. It involves increased transcription of ER chaperones and degradative enzymes.
- uniporter
Carrier protein that transports a single solute from one side of the membrane to the other.
- unsaturated
Describes a molecule that contains one or more double or triple carbon-carbon bonds, such as isoprene or benzene.
- V gene segment
Gene segment encoding most of the variable region of the polypeptide chains of immunoglobulins and T cell receptors.
- V (D) J joining
Recombination process by which gene segments are brought together to form a functional gene for a polypeptide chain of an immunoglobulin or T cell receptor.
- vacuole
Very large fluid-filled vesicle found in most plant and fungal cells, typically occupying more than a third of the cell volume.
- van der Waals attraction
Type of (individually weak) noncovalent bond that is formed at close range between nonpolar atoms.
- variable region
Region of an immunoglobulin light or heavy chain that differs from molecule to molecule; it comprises the antigen-binding site.
- vector
In cell biology, the DNA of an agent (virus or plasmid) used to transmit genetic material to a cell or organism. (See also cloning vector, expression vector.)
- vegetal pole
The end at which most of the yolk is located in an animal egg. The end opposite the animal pole.
- ventral
Situated toward the belly surface of an animal, or towards the underside of a wing or leaf.
- vesicle
Small, membrane-bounded, spherical organelle in the cytoplasm of a eucaryotic cell.
- vesicular transport
Transport of proteins from one cellular compartment to another by means of membrane-bounded intermediaries such as vesicles or organelle fragments.
- virulence gene
Gene that contributes to an organism’s ability to cause disease.
- virus
Particle consisting of nucleic acid (RNA or DNA) enclosed in a protein coat and capable of replicating within a host cell and spreading from cell to cell. Many viruses cause disease.
- voltage-gated cation channel
Type of ion channel found in the membranes of excitable cells (such as nerve cells and muscle) which opens in response to a shift in membrane potential past a threshold value.
- v-SNARE
see SNAREs
- Western blotting
Technique by which proteins are separated by electrophoresis and immobilized on a paper sheet and then analyzed, usually by means of a labeled antibody.
- white blood cell (leucocyte)
General name for all the nucleated blood cells lacking hemoglobin. Includes lymphocytes, neutrophils, eosinophils, basophils, and monocytes.
- wild-type
Normal, nonmutant form of an organism; the form found in nature (in the wild).
- Xenopus laevis (South African clawed toad)
Species of frog (not toad) frequently used in studies of early vertebrate development.
- XIC
- X-inactivation
Inactivation of one copy of the X chromosome in the somatic cells of female mammals.
- X-inactivation center (XIC)
Site in an X chromosome at which inactivation is initiated and spreads outwards.
- X-ray crystallography
Technique for determining the three-dimensional arrangement of atoms in a molecule based on the diffraction pattern of X-rays passing through a crystal of the molecule.
- yeast
Common term for several families of unicellular fungi. Includes species used for brewing beer and making bread, as well as pathogenic species (that is, species that cause disease).
- yolk
Nutritional reserves rich in lipids, proteins and polysaccharides, present in the eggs of many animals.
- Z disc (Z line)
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached. Seen as a dark transverse line in micrographs.
- zinc finger
DNA-binding structural motif present in many gene regulatory proteins. Composed of a loop of polypeptide chain held in a hairpin bend bound to a zinc atom.
- zona pellucida
Glycoprotein layer on the surface of the unfertilized egg. It is often a barrier to fertilization across species.
- zygote
Diploid cell produced by fusion of a male and female gamete. A fertilized egg.
- zygotene
Second stage of division I of meiosis, in which the synaptonemal complex begins to form between the two sets of sister chromatids in each bivalent chromosome.
- Glossary - Molecular Biology of the CellGlossary - Molecular Biology of the Cell
Your browsing activity is empty.
Activity recording is turned off.
See more...