In the preceding chapter, we gave a brief overview of the methods that are commonly used for identification of protein-coding genes and analysis of protein sequences. Here, we turn to one of the main subjects of this book, namely, how these methods are applied to the task of primary analysis of genomes, which often goes under the name of “genome annotation”. Many researchers still view genome annotation as a notoriously unreliable and inaccurate process. There are excellent reasons for this opinion: genome annotation produces a considerable number of errors and some outright ridiculous “identifications” (see 3.1.3 and further discussion in this chapter). These errors are highly visible, even when the error rate is quite low: because of the large numbers of genes in most genomes, the errors are also rather numerous. Some of the problems and challenges faced by genome annotation are an issue of quantity turning into quality: an analysis that can be easily and reliably done by a qualified researcher for one or ten protein sequences becomes difficult and error-prone for the same scientist and much more so for an automated tool when the task is scaled up to 10,000 sequences. We discuss here the performance of manual, automated, and mixed approaches in genome annotation and ways to avoid some common pitfalls. Mostly, however, we concentrate in this chapter on the so-called context methods of genome analysis, which are the recent excitement in the annotation field. These approaches go beyond individual genes and explicitly take advantage of genome comparison.
5.1. Methods, Approaches and Results in Genome Annotation
5.1.1. Genome annotation: data flow and performance
What is genome annotation? Of course, there hardly can be any exact definition but, for the purpose of this discussion, it might be useful to define annotation as a subfield in the general field of genome analysis, which includes more or less anything that can be done with genome sequences by computational means. In simple, operational terms, annotation may be defined as the part of genome analysis that is customarily performed before a genome sequence is deposited in GenBank and described in a published paper. We say “customarily” because the annotations available through GenBank and particularly the types of analysis reported in the literature for different genomes vary widely. For instance, the reports on the human genome sequence [488,870] clearly include a considerable amount of information that goes beyond typical genome annotation. The “unit” of genome annotation is the description of an individual gene and its protein (or RNA) product, and the focal point of each such record is the function assigned to the gene product. The record may also include a brief description of the evidence for this assigned function, e.g. percent identity with a functionally characterized homolog or the boundaries of domains detected in a domain database search, but there is no room for any details of the analysis.
Figure 5.1 shows a rough schematic of the data flow in genome annotation, starting with the finished sequence; we leave finishing of the sequence out of this scheme but indicate the possibility of feedback resulting in correction of sequencing errors. Of these procedures, which must be integrated for predicting gene functions, statistical gene prediction and search of general-purpose databases for sequence similarity are central in the sense that this is done comprehensively as part of any genome project. The contribution of the other approaches in the scheme in Figure 5.1, particularly specialized database search, including domain databases, such as Pfam, SMART, and CDD (see 3.2.2), and genome-oriented databases, such as COGs, KEGG, or WIT (see 3.4), and genomic context analysis, varies greatly from project to project. So far, these relatively new methods and resources remain ancillary to traditional database search in genome annotation, but we argue further in this chapter that they can and probably will transform the annotation process in the nearest future.
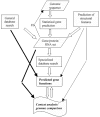
Figure 5.1
A generalized flow chart of genome annotation. FB: feedback from gene identification for correction of sequencing errors, primarily frameshifts. General database search: searching sequence databases (typically, NCBI NR) for sequence similarity, usually (more...)
Before we consider several aspects of genome annotation, it may be instructive to assess its brutto performance, i.e. the fraction of the genes in a genome, to which a specific function is assigned. Table 5.1 lists such data for several genomes sequenced in 2001 and annotated using relatively up-to-date methods. This comparison shows notable differences between the levels of annotation of different genomes. Some genomes simply come practically unannotated, such as, for example, Sulfolobus tokodaii, which is a crenarchaeon closely related to S. solfataricus, and represented in the COGs to the same extent as the latter species. In most genomes, however, functional prediction has been made for the majority of the genes, from 54% to 79% of the protein-coding genes. Obviously, these differences depend both on the taxonomic position of the species in question (e.g. it is likely that for Crenarchaea, whose biology is in general poorly understood, the fraction of genes for which functional prediction is feasible will be lower than for bacteria of the well-characterized Bacillus-Clostridium group, such as C. acetobutylicum or L. lactis) and on the methods and practices of genome annotators.
Table 5.1
Microbial genome annotation 2001.
Even in better-characterized genomes, for hundreds of genes (those encoding “conserved hypothetical” and “hypothetical” proteins), there is no functional prediction whatsoever. Furthermore, among those proteins that formally belong to the annotated category, a substantial fraction of the predictions are only general and are in need of major refinement. Some of these problems can be solved only through experiment, but the above numbers show beyond doubt that there is ample room for improvement in computational annotation itself; further in this chapter, we touch upon some of the possible directions.
Genome annotation necessarily involves some level of automation. No one is going to manually paste each of several thousand-protein sequences encoded in a genome into the BLAST window, hit the button, and wait for the results to appear on screen. For annotation to be practicable at all, software is necessary to run such routine tasks in a batch mode and also to organize the results from different programs in a convenient form, and each genome project employs one or another set of tools to achieve this. After that point, however, genome annotation is still mostly “manual” (or, better, “expert”) because decisions on how to assign gene functions are made by humans (supposedly, experts). Several attempts have been made to push automation beyond straightforward data processing and to allow a program to actually make all the decisions. We briefly discuss some of the automated systems for genome annotation in the next section.
5.1.2. Automation of genome annotation
Terry Gaasterland and Christoph Sensen once estimated that annotating genomic sequence by hand would require as much as one year per person per one megabase [253]. We now believe, on the basis of our own experience of genome annotation (e.g. [622,779,805]), that this estimate is exaggerated perhaps by a factor of 5 or 6. Nevertheless, there is no doubt that genome annotation has become the limiting step in most genome projects. Besides, humans are supposed to be inconsistent and error-prone. Hence the incentives for automating as much of the annotation process as possible.
The GeneQuiz (http://www.sander.ebi.ac.uk/genequiz/) project was the first automatic system for genome analysis, which performed similarity searches followed by automatic evaluation of results and generation of functional annotation by an expert system based on a set of several predefined rules [749]. Several other similar systems have been created since then, but GeneQuiz remains the only such tool that is open to the general public [350].
GeneQuiz runs automated database searches and sequence analysis by taking a protein sequence and comparing it against a non-redundant protein database, generated by automated cross-linking and cross-referencing of PDB, SWISS-PROT, PIR, PROSITE, and TrEMBL databases, with the addition of human, mouse, fruit fly, zebrafish, and Anopheles gambiae protein sets obtained from the Ensemble project (http://www.ensembl.org) and a C. elegans protein set (http://www.sanger.ac.uk/Projects/C_elegans/wormpep). This comparison is done by running BLAST and FASTA programs and is used to identify the cases with high similarity, where function can be predicted. Additionally, searches for PROSITE patterns are performed. Predictions are also made for coiled-coil regions using COILS2 [533], transmembrane segments using PHDhtm [715], and secondary structure elements using PHDsec [718]. The system further clusters proteins from the analyzed genome by sequence similarity [822] and constructs multiple alignments. The results are presented in a table that contains information on the best hits (including gene names, database identifiers, and links to the corresponding databases), predictions for secondary structure, coiled-coil regions, etc. and a reliability score for each item. The functional assignment is then made automatically on the basis of the functions of the homologs found in the database. At this level, functional assignments are qualified as clear or as ambiguous.
The effectiveness and accuracy of such fully automated system have been the subject of a rather heated discussion but still remain uncertain. While the authors originally estimated the accuracy of their functional assignments to be 95% or better [638,749], others reported that only 8 of 21 new functional predictions for M. genitalium proteins made by GeneQuiz could be fully corroborated [466]. A similar discrepancy between the functional predictions made by the GeneQuiz team [31] and those obtained by mostly manual annotation [466] was reported for the proteins encoded in the M. jannaschii genome ([264], see http://www.bioinfo.de/isb/1998/01/0007). It appeared that GeneQuiz analysis suffered from the usual pitfalls of sequence similarity searches (see 3.1.3, the next section and [99,104,264]).
PEDANT, MAGPIE, ERGO, IMAGENE
While GeneQuiz seems to be the only fully automated genome annotation tool that is open to the public for new genome analysis, there have been reports of similar systems developed by other genome annotation groups. These include Dmitrij Frishman's PEDANT (http://pedant.gsf.de, [245,248], Terry Gaasterland's MAGPIE and its sister programs (http://genomes.rockefeller.edu, [252,253]), Ross Overbeek's ERGO (http://ergo.integratedgenomics.com/ERGO, [642,643]), Alan Viari's Imagene (http://wwwabi.snv.jussieu.fr/research, [561]), and some others. Although none of these systems is freely available to outside users, many of the genome annotation results they produced are accessible on the web and can be used to judge the performance.
The PEDANT web site contains by far the most information open to the public and can be used as a good reference point for automated genome analyses (see also 2.4).
SEALS
In addition to completely automated systems, some tools that greatly facilitate and accelerate manual genome annotation are worth a mention. System for Easy Analysis of Lots of Sequences (SEALS), developed by Roland Walker at the NCBI is, for obvious reasons, the one most familiar to the authors of this book (available for downloading at http://iubio.bio.indiana.edu:7780/archive/00000466/, [878]). The SEALS package consists of ~50 simple, UNIX-based tools (written in PERL), which follow consistent syntax and semantics. SEALS combines software for retrieving sequence information, scripting database searches with BLAST, viewing and parsing search outputs, searching for protein sequence motifs using regular expressions, and predicting protein structural features and motifs. Typically, using SEALS, a genome analyst first looks for structural features of proteins, such as signal peptides (predicted by SignalP), transmembrane domains (predicted by PHDhtm), coiled-coil domains (predicted by COILS2), and large non-globular domains (predicted using SEG). Once these regions are identified and masked, database searches are run in a batch mode using the chosen method, e.g. PSI-BLAST. The outputs can be presented in a variety of formats, of which filtering with taxonomic queries implemented in the SEALS script TAX_COLLECTOR is among the most useful. SEALS has been extensively used in the comparative studies of bacterial, archaeal, and eukaryotic genomes (e.g. [52,55,540].
5.1.3. Accuracy of genome annotation, sources of errors, and some thoughts on possible improvements
Benchmarking the accuracy of genome annotation is extremely hard. It has been shown on numerous occasions that more advanced methods for sequence comparison, such as gapped BLAST and subsequently PSI-BLAST, sometimes used in combination with threading, as well as various forms of motif analysis and careful manual integration of the results produced by all these approaches, substantially improve detection of homologs (e.g. [168,401,434,466,585]). At the end, however, genome annotation is not about detection of homologs but rather about functional prediction, and here, the problem of a standard of truth is formidable. By definition, functional annotation (more precisely, functional prediction) deals with proteins whose functions are unknown, and the rate of experimental testing of predictions is extremely slow. We believe that it is possible to design an objective test of the accuracy of genome annotation in the following manner. The protein set encoded in a newly sequenced genome is analyzed, and specific active centers and other functionally important sites are predicted for as many proteins as possible. When a new, preferably phylogenetically distant genome becomes available, orthologs of the proteins from the first genome are identified, and the conservation of the predicted functional sites is assessed. Lack of conservation would count as an error; this is, of course, a harsh test that would give the low bound of accuracy because: first, functional site prediction may be partly wrong but the function of the protein still would be predicted correctly; and second, some active sites might be disrupted in the new genome. In this way, the accuracy of the prediction could be assessed quantitatively and, in principle, even a “tournament” analogous to the CASP competition in protein structure prediction [869] could be arranged.
However, so far, evaluation of the accuracy of genome annotation has been largely limited to the assessments of consistency of annotations of the same genome generated by different groups and various “sanity checks” and expert judgments. Steven Brenner published an interesting comparison of three independent annotations [242,467,639] of the smallest of the sequenced bacterial genomes, Mycoplasma genitalium [116]. Without attempting to determine which annotation was “better”, he manually examined all conflicting annotations, eliminating trivial semantic differences and counting the apparent irreconcilable ones as errors (in at least one of the annotations). His conclusion was that there was an at least 8% error rate among the 340 genes annotated by at least two of the three groups. In a similar exercise that we have done on the basis of the COG database, we found that of 786 COGs that did not include paralogs (the number for the end of 1999), members of 194 had conflicting annotations in GenBank [267]. This suggests, more pessimistically, an annotation error rate of at least 25% using the same criterion as applied by Brenner. Clearly, even the lower of these estimates represents a serious problem for genome annotation, bringing up the specter of error catastrophe [89,104]. We first briefly discuss the most common sources of errors and then some ideas regarding the ways out. Manual and automated genome annotation encounter the same typical problems, which we already mentioned in the discussion of the reliability of sequence database records (see 3.1.3). Inevitably, even partial automation of the annotation process tends to increase the likelihood of all these types of errors.
In order to examine various kinds of errors that are common in genome annotation, it is convenient to re-examine four cases of discrepancies in the annotation of M. genitalium proteins that were specifically highlighted in the aforecited article of Steven Brenner (Table 5.2). Although one of the authors was involved in one of the compared annotations, we think we can be completely impartial in the spirit of Brenner's article, especially since six years have passed, an eternity for genomics.
Table 5.2
Different types of errors in genome annotation.
The protein MG302 was not annotated in the original genome publication by Fraser and colleagues and was assigned conflicting annotations by the other two groups. Ouzounis and coworkers notably characterized this protein as a “mitochondrial 60S ribosomal protein L2”, whereas Koonin and coworkers annotated it is as a permease, perhaps specific for glycerol-3-phosphate. A database search performed in 2002 leaves no doubt whatsoever that the protein is a permease; this is, of course, readily supported by transmembrane segment prediction. However, the glycerol-3-phosphate specificity is not supported at all. Instead, these searches, particularly the CDD search, unequivocally pointed to a relationship between MG302 and a family of cobalt transporters. Nevertheless, since the similarity between MG302 and the cobalt transporters is not particularly strong and transporters switch their specificity with relative ease during evolution, caution is due, and the annotation as “probable Co transporter” seems most appropriate. This single case nicely covers several common problems of genome annotation. The most benign but also apparently most widespread of these is overprediction or, more precisely, overly specific prediction . Even with the methods available in 1996 (ungapped BLAST, FASTA, various alignment methods, and transmembrane segment prediction), the conclusion that MG302 was a permease was quite firm. However, glycerol-3-phosphate permease turned up as the most similar functionally characterized protein just by chance (Co2+ transporters had not been characterized at the time). Transferring functional information from this unreliable best hit, however tentatively, was a typical error of overprediction; the appropriate annotation at the time would have been, simply, “predicted permease”. The annotation of MG302 as “mitochondrial 60S ribosomal protein L2” is, of course, much more conspicuous. At face value, this does not even pass a “reality check”: there certainly can be no mitochondria and no 60S ribosomes in mycoplasmas.
Such semantic snafus are pretty common in genome annotation, especially those that are either produced fully automatically or manually but non-critically (e.g. the “discovery” of head morphogenesis in bacteria mentioned in Chapter 3). However, these are probably the least serious annotation errors.
Let us just assume that the authors of this annotation meant “homolog of mitochondrial 60S ribosomal protein L2”. What is worse: the search result that presumably gave rise to this annotation is impossible to reproduce at this time, at least not without detailed research, which we are not willing to undertake. It is most likely that this blatantly wrong annotation was due to a spurious database hit to a ribosomal protein that was not critically assessed. It is not clear, in this particular case, how could this spurious hit pass the significance threshold, but in general, this happens most often because of the lack of proper filtering for low complexity (or alternative approaches, such as composition-based statistics, which are available in 2002 but had not been developed in 1996; see Chapter 4). Alternatively or additionally, the problem might lie in non-critical transfer of annotation from an unreliable database record , i.e. a low-complexity sequence erroneously labeled as a ribosomal protein. Notably, our re-analysis shows that the annotations assigned by each of the three groups were not completely correct: one was an outright error; another one involved overprediction; and the third one, an underprediction. Although less notorious than false predictions (false-positives, in statistical terms), lack of prediction, where a confident one is feasible with available methods, is still an error (a false-negative).
The case of the MG225 protein is quite similar except that there was no clear false prediction involved. Once again, the original genome project gave no annotation (a false-negative), whereas one of the remaining groups annotated the protein as “histidine permease”, and the other one stopped at an “amino acid permease” annotation without proposing specificity. Today's searches support the latter decision because no convincing, specific relationship between this protein and transporters for any particular amino acid could be detected (in fact, given the small repertoire of transporters in mycoplasmas, this one might have a broad specificity). Notably, both MG302 and MG225 remain “hypothetical proteins” in GenBank to this day, although closely related orthologs from M. pneumoniae are correctly annotated as permeases [168].
The MG085 protein was annotated as an oxidoreductase (of different families) in the original genome report and by Ouzounis and coworkers, whereas Koonin and coworkers predicted that it was an ATP(GTP?)-utilizing enzyme on the basis of the conservation of the P-loop motif in this protein and its homologs. In 2002, database searches immediately identify this protein as HPr kinase (this annotation is now correctly assigned to MG085 in GenBank), a regulator of the sugar phosphotransferase system, which indeed is a P-loop-containing, ATP-utilizing enzyme [723]. Back in 1996, this was the only informative annotation that could be derived for this protein; HPr kinase genes had not been identified at the time. Once again, the specific source of the oxidoreductase assignments is hard to determine; spurious hits, non-critical use of incorrect database annotations, or a combination thereof must have caused this.
The case of MG448 is of particular interest. This protein was annotated as “pilin repressor” or simply PilB protein by Fraser and coworkers and Ouzounis and coworkers and, somewhat cryptically, as “chaperone-like protein” by Koonin and coworkers. This protein remains “hypothetical” in GenBank but became a peptide methionine sulfoxide reductase (PMSR) in SWISS-PROT. A database search detects highly significantly similarity with numerous proteins that are annotated primarily as PMSR and, in some cases, as PilB-related repressors. In reality, this protein is indeed a recently characterized, distinct form of PMSR, MsrB [476,526], which is evolutionarily unrelated to, but is often associated with, the classic PMSR, MsrA, either as part of a multidomain protein or as a separate gene in the same operon [267]. These fusions resulted in the annotation of MG448 as PMSR, which, ironically, turned out to be correct, but mostly (except for the recently updated SWISS-PROT description), for a wrong reason, because it was the MsrA domain that was recognized in the fusion proteins. Furthermore, in several bacteria, these two domains are fused to a third, thioredoxin domain. The three-domain protein of Neisseria gonorrhoeae has been characterized as a regulator of pili operon expression, and this is what caused the annotation of MG448 as PilB, which was reproduced by two groups. This annotation is outright wrong and does not even pass a “reality check” because there are no pili in mycoplasmas (parenthetically, latest reports appear to indicate that even the original functional characterization of the Neisseria protein was erroneous [776]).
Unrecognized multidomain architecture of either the analyzed protein or its homologs or both is a common cause of erroneous annotation. The “chaperone-like protein” annotation was based on the notion that the PMSR function could be interpreted as a form of chaperone action, and accordingly, the associated domain was also likely to have a chaperone-like activity. In retrospect, this looks like overprediction combined with insufficient information included in the annotation. A straightforward annotation of MG448 as a PMSR-associated domain, perhaps with an extra prediction of redox activity on the basis of conservation of cysteines in this domain, the way it has been done in a subsequent publication [267], would have been appropriate. We revisit this interesting set of proteins when discussing context analysis in Section 5.2.
While considering only four proteins with contradictory annotations, we encountered all the main sources of systematic error in genome annotation. We list them here again, more or less in the order of decreasing severity, as we see it: (i) spurious database hits, often caused by low-complexity regions in the query or the database sequence; (ii) non-critical transfer of functional prediction from an unreliable database record; (iii) incorrect interpretation (lack of recognition) of multidomain architecture of the query and/database sequences; (iv) overly specific functional prediction; and (v) underprediction.
We believe that this brief discussion highlights more general problems beyond these specific causes of errors. Even the apparently correct database annotations are insufficiently informative. Typically, the records do not include the evidence behind the prediction or include only minimal data that may be hard to interpret, such as E-values of the hits to particular domains. In this situation, any complicated case will not be represented adequately (e.g. the PMSR-associated domain discussed above). In addition, there is no controlled vocabulary for genome annotation, which creates numerous semantic problems, although an attempt to correct this situation is being undertaken in the form of the Genome Ontology project [60,513].
The above discussion shows that the general state of genome annotation is far from being satisfactory. What can be done to improve it? In his paper on genome annotation errors, Steven Brenner noted that, “to prevent errors from spreading out of control, database curation by the scientific community will be essential.” [116]. Curation, however, implies that databases other than GenBank will have to be employed because GenBank, by definition, is an archival database (Chapter 3). It appears that the future and, to some degree, already the present of genome annotation lies in specialized databases that actually function as annotation tools. The beginnings of such tools can be seen in databases like KEGG, WIT, and COGs, complemented by tools for domain identification, such as CDD and SMART (see Chapters 3 and 4).
Conceptually, the advantage of this approach may be viewed as reduction and structuring of the search space for genome annotation. Thus, when using COGs, a genome analyst compares each protein sequence not to the unstructured set of more than a million proteins (the NR database) but instead to a collection of ~5,000 mostly well-characterized protein sets classified by orthology, which is the appropriate level of granularity for functional assignment. Already genome annotation today is starting to change through the use of the new generation of databases and tools. However, smooth integration of these and development of new, richer formats for annotation are things of the future. In the next subsection, we turn to a specific example to illustrate how the use of COGs helps genome annotation.
5.1.4. A case study on genome annotation: the crenarchaeon Aeropyrum pernix
Aeropyrum pernix was the first representative of the Crenarchaeota (one of the two major branches of archaea; see Chapter 6) and the first aerobic archaeon whose genome has been sequenced [427]. A. pernix was reported to encode 2,694 putative proteins in a 1.67-Mbase genome. Of these, 633 proteins were assigned a specific or general function in the original report on the basis of sequence comparison to proteins in the GenBank, SWISS-PROT, EMBL, PIR, and Owl databases. Given the intrinsic interest of the first crenarchaeal genome and also because of the unexpectedly low fraction of predicted genes that were assigned functions in the original report, A. pernix was chosen for a pilot annotation project centered around the COG database [605].
Figure 5.2 (see the color plates) shows the protocol employed for the COG-based genome annotation. This procedure was not limited to straightforward COGNITOR analysis but also explicitly drew from the phyletic patterns. Whenever A. pernix was unexpectedly not represented in a COG (e.g. a COG that included all other archaeal species), additional analysis was undertaken. To identify possible diverged COG members from A. pernix, PSI-BLAST searches were run with multiple members of the respective COGs, and to detect COG members that could have been missed in the original genome annotation, the translated sequence of the A. pernix genome was searched using TBLASTN. Conversely, unexpected occurrence of A. pernix proteins in COGs that did not have any other archaeal members were examined case by case to detect likely HGT events and novel functions in the crenarchaeal genome.

Figure 5.2
Protocol of genome annotation using the COG database.
Proteins were assigned to COGs through two rounds of automated comparison using COGNITOR, each followed by curation, that is, manual checking of the assignments. The first round attempts to assign proteins to existing COGs; typically, >90% of the assignments are made in this step. The second round serves two purposes: first, to assign paralogs, that might have been missed in the first round, to existing COGs; and, second, to create new COGs from unassigned proteins.
The results of COG assignment for A. pernix are shown in Table 5.3. Manual curation of the automatic assignments revealed a false-positive rate of less than 2% (23 of 1123 proteins). Even if the less severe errors, when a protein was transferred from one related COG to another, are taken into account, the false-positive rate was 4%, which is not negligible but substantially lower than the estimates cited above for more standard genome annotation methods. The number of identified false-negatives was even lower, but in this case, of course, it is not possible to determine how many proteins remain unassigned. It is further notable that the great majority of assigned proteins belonged to pre-existing COGs, which facilitates a (nearly) automatic annotation.
Table 5.3
Assignment of predicted Aeropyrum pernix proteins to COGs.
Altogether, 1,102 A. pernix proteins were assigned to COGs. Some of these proteins (154) were members of functionally uncharacterized COGs. Subtracting these, annotation has been added to 315 proteins, which is an increase of about 50% compared to the original annotation. These newly annotated A. pernix proteins included, among others, the key glycolytic enzymes glucose-6-phosphate isomerase (APE0768, COG0166) and triose phosphate isomerase (APE1538, COG0149), and the pyrimidine biosynthetic enzymes orotidine-5′-phosphate decarboxylase (APE2348, COG0284), uridylate kinase (APE0401, COG0528), cytidylate kinase (APE0978, COG1102), and thymidylate kinase (APE2090, COG0125). Similarly, important functions in DNA replication and repair were confidently assigned to a considerable number of A. pernix proteins, which, in the original annotation, were described as “hypothetical”. Examples include the bacterial-type DNA primase (COG0358), the large subunit of the archaeal-eukaryotic-type primase (COG2219), a second ATP-dependent DNA ligase (COG1423), three paralogous photolyases (COG1533), and several helicases and nucleases of different specificities.
The case of the large subunit of the archaeal-eukaryotic primase is particularly illustrative of the contribution of different types of inference to genome annotation. COGNITOR failed to assign an A. pernix protein to the respective COG (COG2219). However, given the ubiquity of this subunit in euryarchaea and eukaryotes and the presence of a readily detectable small primase subunit in A. pernix (COG1467), a more detailed analysis was undertaken by running PSI-BLAST searches against the NR database with all members of COG2219 as queries. When the A. fulgidus primase sequence (AF0336) was used to initiate the search, the A. pernix counterpart (APE0667) was indeed detected at a statistically significant level.
An interesting case of re-annotation of a protein with a critical function, which also led to more general conclusions, is the archaeal uracil DNA glycosylase (UDG; COG1573). The members of this COG were originally annotated (and still remain so labeled in GenBank) as a “DNA polymerase homologous protein” (APE0427 from A. pernix) or as a “DNA polymerase, bacteriophage type” (AF2277 from A. fulgidus) or as a hypothetical protein. However, UDG activity has been experimentally demonstrated for the COG1573 members from T. maritima and A. fulgidus [740,741]. The reason for the erroneous annotation of these proteins as DNA polymerases is already well familiar to us: independent fusion of the uracil DNA glycosylase with DNA polymerases was detected in bacteriophage SPO1 and in Yersinia pestis [44]. Although these fusions hampered the correct annotation in the original analysis of the archaeal genomes, they seem to be functionally informative, suggesting that this type of UDG functions in conjunction with the replicative DNA polymerase.
The 1,102 COG members from A. pernix comprise 41% of the total number of predicted genes. This percentage was significantly lower than the average fraction of COG members (72%) for the other archaeal species. It seems most likely that this was due to an overestimate of the total number of ORFs in the genome. Many of the A. pernix ORFs with no similarity to proteins in sequence databases (1,538, or 57.1%) overlap with ORFs from conserved families, including COG members. On the basis of the average representation of all genomes in the COGs (67%) and the average for the other archaea (72%), one could estimate the total number of A. pernix proteins to be between 1,550 and 1,700. This range is also consistent with the size of the A. pernix genome (1.67 Mb), given the gene density of about one gene per kilobase, which is typical of bacteria and archaea. More conservatively, 849 ORFs, originally annotated as probable protein-coding genes, significantly overlapped with COG members and could be confidently eliminated, which brings the total number of protein-coding genes in A. pernix to a maximum of 1,873. Unfortunately, the spurious ORFs still remain in the NR database, polluting it and potentially even leading to the emergence of ghost “protein” families once new, related genomes are sequenced. Evidence has been presented that spurious “proteins” have been produced by other microbial genome products also [777], although probably not on the same scale as A. pernix. This regrettable pollution emphasizes the value of specialized, curated databases that are free of apparitions.
Despite this overrepresentation of ORFs in A. pernix, we nonetheless added 28 previously unidentified ORFs that were detected by searching the genome sequence translated in all six frames for possible members of COGs with unexpected phyletic patterns. These newly detected genes represent conserved protein families, including functionally indispensable proteins, such as chorismate mutase (APE0563a, COG1605), translation initiation factor IF-1 (APE_IF-1, COG0361), and seven ribosomal proteins (APE_rpl21E, COG2139; APE_rps14, COG0199; APE_rpl29, COG0255; APE_rplX, COG2157; APE_rpl39E, COG2167; APE_rpl34E, COG2174; APE_rps27AE, COG1998).
This pilot analysis, while falling far short of the goal of comprehensive genome annotation, highlights some advantages of specialized comparative-genomic databases as annotation tools. In this particular case, the original annotation probably had been overly conservative, which partly accounts for the large increase in the functional prediction rate. However, the employed protocol is general and, with modifications and addition of some extra procedures, has been used in primary genome analysis [622,779]. In other genome projects, the WIT system has been employed in a conceptually similar manner [179,418]. As shown above, this type of analysis yields reasonable accuracy of annotation, even when applied in a fully automated mode (Table 5.3). However, additional expert contribution, particularly in the form of context analysis discussed in the next section, adds substantial value to genome annotation.
5.2. Genome Context Analysis and Functional Prediction
All the preceding discussion in this chapter centered on prediction of the functions of proteins encoded in sequenced genomes by extrapolating from the functions of their experimentally characterized homologs. The success of this approach depends on the sensitivity and selectivity of the methods that are used for detecting sequence similarity (see Chapter 4) and on the employed rules of inference (see 5.1). There is no doubt that homology analysis remains the central methodology of genomics, i.e. the one that produces the bulk of useful information. However, a group of recently developed approaches in comparative genomics goes beyond sequence or structure comparison. These methods have become collectively and, we think, aptly known as genome context analysis [267,368,369,372]. The notion of “context” here includes all types of associations between genes and proteins in the same or in different genomes that may point to functional interactions and justify a verdict of “guilt by association” [36]: if gene A is involved in function X and we obtain evidence that gene B functionally associates with A, then B is also involved in X. More specifically, context in comparative genomics pertains to phyletic profiles of protein families, domain fusions in multidomain proteins, gene adjacency in genomes, and expression patterns. Indeed, genes whose products are involved in closely related functions (e.g. form different subunits of a multisubunit enzyme or participate in the same pathway) should all be either present or absent in a certain set of genomes (i.e. have similar if not identical phyletic patterns) and should be coordinately expressed (i.e. are expected to be encoded in the same operon or at least to have similar expression patterns). This simple logic gives us a potentially powerful way to assign genes that have no experimentally characterized homologs to particular pathways or cellular systems. Although context methods usually provide only rather general predictions, they represent a new and important development in genomics that explicitly takes advantage of the rapidly growing collection of sequenced genomes.
5.2.1. Phyletic patterns (profiles)
Genes coding for proteins that function in the same cellular system or pathway tend to have similar phyletic patterns [259,828]. Numerous examples for a variety of metabolic pathways are given in Chapter 7. These observations led to the suggestion that this trend could be used in the reverse direction, i.e. to deduce functions of uncharacterized genes [665]. However attractive this idea might be, the real-life phyletic patterns are heavily affected by such major evolutionary phenomena as partial redundancy in gene functions, non-orthologous gene displacement, and lineage-specific gene loss. As a result, there are thousands different phyletic patterns in the COGs, most of them represented only once or twice. Moreover, examination of a variety of multi-component systems and biochemical pathways (http://www.ncbi.nlm.nih.gov/cgi-bin/COG/palox?sys=all) shows that, despite the tendency of the components of the same complex or pathway to have similar patterns, there is not even one pathway in which all members show exactly the same pattern. Even the principal metabolic pathways, such as glycolysis, TCA cycle, and purine and pyrimidine biosynthesis, show considerable variability of phyletic patterns due to non-orthologous gene displacement ([265,270,370], see Chapter 7).
Because of this variability, the predictive power of the observation that two genes have the same phyletic pattern is, in and by itself, limited. However, when supported by other lines of evidence, such observations prove useful. Somewhat counterintuitively, the universal pattern is one of the most strongly indicative of gene function: among the 63 universal COGs, at least 56 consist of proteins involved in translation. The functions of those few proteins in the universal set that remain uncharacterized can be predicted with considerable confidence through combination of this phyletic pattern with other lines of evidence. For example, the uncharacterized protein YchF, which belongs to the universal set (COG0012), is predicted by sequence analysis to be a GTPase; in addition, this protein contains a C-terminal RNA-binding TGS domain [909]. Taken together with the ubiquity of this protein and with the fact that, in phylogenetic trees, the archaeal members of the COG clearly cluster with eukaryotic ones, this strongly suggests that YchF is an uncharacterized, universal translation factor [267]. This is supported by the juxtaposition of the ychF gene with the gene for peptidyl-tRNA hydrolase (pth) in numerous proteobacteria. The discussion of this protein made us run ahead of ourselves and invoke other context methods, which are considered in the next subsections, namely, analysis of domain fusions and gene juxtaposition. This situation is quite typical: context methods are at their best when they complement one another. Although statistical significance estimates for a combination of context methods do not currently seem feasible, in a case like YchF, the evidence appears to be, for all practical purposes, irrefutable.
Another similar case involves the predicted ATPase or (more likely) kinase YjeE from E. coli [256] and its orthologs from a majority of bacterial genomes that comprise COG0802. Domain analysis identified this protein as a likely P-loop ATPase but failed to give any indications as to its cellular role. The phyletic pattern of this COG shows that YjeE is encoded in every bacterial genome, with the exception of M. genitalum, M. pneumoniae, and U. urealyticum, the only three bacterial species in the COG database that do not form a cell wall. Since other conserved proteins with the same phyletic pattern (MurA, MurB, MurG, FtsI, FtsW, DdlA) are enzymes of cell wall biosynthesis, it can be predicted that YjeE is an ATPase or kinase involved in the same process. Again, this prediction is supported by the adjacency of the yjeE with the gene for N-acetylmuramoyl-L-alanine amidase, another cell wall biosynthesis enzyme.
There is more to phyletic pattern analysis then prediction based on identical or similar patterns. Guilt by association can be established also through identification of sets of genes that are co-eliminated in a given lineage; this approach exploits the widespread phenomenon of lineage-specific gene loss. A systematic analysis of the set of genes that have been co-eliminated in the yeast S. cerevisiae after its divergence from the common ancestor with S. pombe led to the prediction that a particular group of proteins, including one that contained a helicase and a duplicated RNAse III domain, was involved in post-transcriptional gene silencing [55]. This protein turned out to be the now famous dicer nuclease, which indeed has a central role in silencing [365,436].
On many occasions, non-orthologous gene displacement manifests in complementary , rather than identical or similar, phyletic patterns, like we have seen for phosphoglycerate mutase in 2.2.6. The complementarity is rarely perfect because of partial functional redundancy: some organisms, particularly those with larger genomes, often encode more than one protein to perform the same function. This can be illustrated by the case of the recently discovered new type of fructose-1,6-bisphosphate aldolase, referred to as FbaB or DhnA [257]. The two well-known variants of this enzyme, class I (Schiff-base forming, metal-independent) and class II (metal-dependent), have long been considered to be unrelated (analogous) enzymes until structural comparisons revealed their underlying similarity (see Figure 1.9) [95,187,257,549]. These enzymes are generally limited in their phyletic distribution to eukaryotes (class I) and bacteria (class II); some bacteria, however, have both variants and yeast has the bacterial (class II) form of the enzyme [549]:
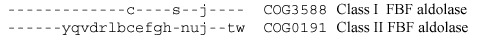
Sequencing of archaeal genomes revealed the absence of either form of the fructose-1,6-bisphosphate aldolase. The same was the case with chlamydiae, which were predicted to have a third form of this enzyme [412,805]. Indeed, investigation of the metal-independent fructose-1,6-bisphosphate aldolase activity in E. coli led to the discovery of another metal-independent Schiff-base-forming variant [844] whose sequence, however, was more closely related to those of class II enzymes than to typical class I enzymes [257]. Highly conserved homologs of this new, third form of fructose-1,6-bisphosphate aldolase were found in chlamydial and archaeal genomes:

As with phosphoglycerate mutase, combining these phyletic patterns shows almost perfect complementarity, with aldolase missing only in Rickettsia, which does not encode any glycolytic enzymes, and in Thermoplasma, which appears to rely exclusively on the Entner-Doudoroff pathway (see 7.1.1):

Other interesting examples of complementary phylogenetic patterns include lysyl-tRNA synthetases, pyridoxine biosynthesis proteins PdxA and PdxZ [256], thymidylate synthases [267], and many others. The case of thymidylate synthases is particularly remarkable. Thymidylate synthase is a strictly essential enzyme of DNA precursor biosynthesis, and its apparent absence in several bacterial and archaeal species became a major puzzle as their genome sequences were reported.
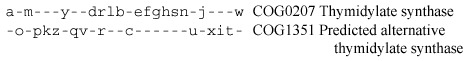
The alternative thymidylate synthase was predicted [267] on the basis of a phyletic pattern that was nearly complementary (with just one case of redundancy) to that of the classic thymidylate synthase (ThyA) and the report that the homolog of the COG1351 proteins from Dictyostelium complemented thymidylate synthase deficiency [206]. Just before this book went to print, a new issue of Science reported the confirmation of this prediction: not only was it shown that the COG1351 member from H. pylori had thymidylate synthase activity, but also the structure of this proteins has been solved and turned out to be unrelated to that of ThyA [589,598].
5.2.2. Gene (domain) fusions: “guilt by association”
It is fairly common that functionally interacting proteins that are encoded by separate genes in some organisms are fused in a single polypeptide chain in others. This has been confirmed by statistical analysis that demonstrated general functional coherence of fused domains [930]. The advantages of a multidomain architecture are that this organization facilitates functional complex assembly and may also allow reaction intermediate channeling [546].
The basic assumption in the analysis of domain fusions is that a fusion will be fixed during evolution only when it provides a selective advantage to the organism in the form of improved functional interaction between proteins. Thus, finding fused proteins (domains) in one species suggests that they might interact, physically or at least functionally, in other species. In and by itself, this notion is trivial and has been employed for predicting protein and domain functions on an anecdotal basis for years (see [100], just as an example). However, with the rapid growth of the sequence information, the applicability of this approach widened and two independent groups proposed, in well-publicized papers, that analysis of domain fusions could be a general method for systematic and, moreover, automatic, prediction of protein functions [213,546]. In one of these studies [546], domain fusions are referred to as “Rosetta Stone” proteins – clues to deciphering the functions of their component domains, and this memorable name stuck to the whole approach. (The Rosetta Stone metaphor is quite loose: the notorious stone used by François Champollion to decipher the Egyptian hieroglyphs and now on public display in the British Museum, is a tri-lingua, i.e. a monument that has on it the same text in three different languages. There is nothing exactly like that about domain fusions, it is just possible to say vaguely that the “language” of domain fusions is translated into the “language” of functional interactions. The “guilt by association” simile [36] seems much more apt if less glamorous).
In his comment on the “Rosetta Stone” excitement, Russell Doolittle pointed out that cases that establish a link between two well-known domains or those that link two unknown domains are not likely to lead to any scientific breakthroughs [188]. Only those “Rosetta Stone” proteins, in which an unknown domain is linked to a previously characterized one, can be used to infer the function(s) of the uncharacterized domain. Analysis of domain fusions in complete microbial genomes indicates that they are a complex mixture of informative, uninformative and potentially misleading cases, which certainly provide many clues to functions of uncharacterized domains. However, interpretations stemming from domain fusion seem to require case-by-case examination by human experts and, most of the time, become really useful only when combined with other lines of evidence.
One of the advantages of the guilt by association approach is that, at least in principle, it allows transitive closure, i.e. expansion of functional associations between transitively connected components. In other words, detection of domain combinations AB, BC, and CD suggests that domains A, B, C and D form a functional network. This approach has been successfully applied to the analysis of prokaryotic signal-transduction systems, resulting in the prediction of several new signaling domains. Participation of these domains in signaling cascades has been originally proposed solely on the basis of their conserved domain architectures and subsequently confirmed experimentally [269].
In Figure 5.3, we illustrate the “guilt by association” approach using the peptide methionine sulfoxide reductase example discussed in the previous section as a case of annotation complicated by domain fusion. As in the examples above, the logic of the analysis does not allow us to use domain fusions only; we also have to invoke phyletic patterns and organization of genes in the genome.

Figure 5.3
A Rosetta Stone case: domain fusions and gene clusters that involve peptide methionine sulfoxide reductases.
In most organisms, protein methionine sulfoxide reductase A (MsrA) is a small, single-domain protein. However, in H. influenzae, H. pylori and T. pallidum, it is fused with another, highly conserved domain (MsrB) that is found as a distinct protein in all other organisms that encode MsrA. In other words, the two fusion components show the same phyletic patterns:

In B. subtilis, the genes for MsrA and MsrB are not fused, but are adjacent and may form an operon. In contrast, in T. pallidum, MsrA and MsrB are fused, but in reverse order, compared to H. influenzae and H. pylori (Figure 5.3). The H. influenzae and H. pylori “Rosetta Stone” proteins are most closely related to each other, but the one from T. pallidum does not show particularly strong similarity to any of them, suggesting two independent fusion events in these two lineages.
In Neisseria and Fusobacterium, a third, thioredoxin-like domain joins the MsrAB fusion (Figure 5.3). In H. influenzae, the ortholog of this predicted thioredoxin is encoded two genes upstream of MsrAB. The gene in between encodes a conserved integral membrane protein, designated CcdA for its requirement for cytochrome c biogenesis in B. subtilis. Its ortholog is encoded next to MsrAB in H. pylori and next to thioredoxin in several other genomes (Figure 5.3).
Combining all this evidence from the guilt by association approach, gene adjacency data, phyletic profiles, and sequence analysis, it has been predicted that the MsrA, MsrB and thioredoxin form an enzymatic complex, which catalyzes a cascade of redox reactions and is associated with the bacterial membrane via CcdA. However, this is probably not the only complex in which MsrAB is involved, because not all genomes that have this gene pair also encode CcdA (Figure 5.3). Since the publication of this prediction, it has been largely confirmed by the demonstration that MsrB is a second, distinct, thioredoxin-dependent peptide methionine sulfoxide reductase, which cooperates with MsrA in the defense of bacterial cells against reactive oxygen species [316,526,776]. However, the CcdA connection remains to be investigated.
This case study demonstrates both the considerable potential of domain fusion analysis as a tool for protein function prediction, particularly when combined with other context-based and homology-based approaches, and potential problems. One could be tempted to extend the small network of domains shown in Figure 5.3 by including other domains that form fusions (or are encoded by adjacent genes) with the thioredoxin domain. It appears, however, that such an extension would have been ill-advised. Firstly, orthologous relationships among thioredoxins are ambiguous, and secondly, although thioredoxins are not among the most “promiscuous” domains, the variety of their “guilt by association” links still is sufficiently large to make any predictions regarding potential functional connections between the respective domains and MsrAB dubious at best. These two issues, identification of orthologs and “promiscuity” characteristic of certain domains, are the principal problems encountered by the “guilt by association” approach. Domain fusions often are found only within a specialized, narrow group of orthologous protein domains, and translating their functional interaction into a general prediction for the respective domains is likely to be grossly misleading. A relatively small number of “promiscuous” domains, particularly those involved in signal transduction and different forms of regulation (e.g. CBS, PAS, GAF domains), combine with a variety of other domains that otherwise have nothing in common and therefore significantly increase the number of false-positives among the Rosetta Stone predictions. Although it is possible to simply exclude the worst known offenders from any Rosetta Stone analysis [546], other domains also have the potential of showing “illicit” behavior and compromising the results. Manual detection of such cases is relatively straightforward, but automation of this process may be complicated.
5.2.3. Gene clusters and genomic neighborhoods
As already mentioned in Chapter 2, comparisons of complete bacterial genomes have revealed the lack of large-scale conservation of the gene order even between relatively close species, such as E. coli and H. influenzae [595,829] or E. coli and P. aeruginosa (Figure 2.6B). Although these pairs of genomes have numerous similar strings of adjacent genes (most of them predicted operons), comparisons of more distantly related bacterial and archaeal genomes have shown that, at large phylogenetic distances, even most of the operons are extensively rearranged [461,884]. The few operons that are conserved across distantly related genomes typically encode physically interacting proteins, such as ribosomal proteins or subunits of the H-ATPase and ABC-type transporter complexes [169,385,461,595].
It should be noted that only a relatively small number of operons have been identified experimentally, primarily in well-characterized bacteria, such as E. coli and B. subtilis [363,732]. However, analysis of gene strings that are conserved in bacterial and archaeal genome strongly suggested that the great majority of them do form operons [916]. This conclusion was based on the following principal arguments: (i) as shown by Monte Carlo simulations, the likelihood that identical strings of more than two genes are found by chance in more than two genomes is extremely low; (ii) most of those conserved strings that include characterized genes either are known operons or include functionally linked genes and can be predicted to form operons; (iii) typical conserved gene strings include 2 to 4 genes, which is the characteristic size of operons; (iv) conserved gene strings that include genes from adjacent, independent operons are extremely rare; (v) nearly all conserved gene strings consist of genes that are transcribed in the same direction [916]. As a result, one can usually assume that conserved gene strings are co-regulated, i.e. form operons, even if they contain additional promoters.
Pairwise genome comparisons showed that, on average, ~10% of the genes in each genome belong to gene strings that are conserved in at least one of the other available genomes [385,916]. These numbers vary widely from <5% for the cyanobacterium Synechocystis sp. to 23–24% in T. maritima and M. genitalium; the fraction of genes that belonged to predicted operons in the archaeal genomes was only slightly lower than that in bacterial genomes [916].
These observations indicate that conserved gene strings are under stabilizing selection that prevents their disruption. For functionally related genes (e.g. those encoding proteins that function in the same pathway or multimeric complex), this selective pressure probably comes from the necessity to synchronize their expression. This conclusion holds even in the face of the “selfish operon” hypothesis, which posits that operons survive during evolution because they are disseminated via HGT [494,495]. We believe that the selfish operon hypothesis seems to put the cart ahead of the horse: operons certainly do spread via HGT, but their transfer leads to fixation more often than transfer of individual genes because of the selective advantage conferred to the recipient by the acquired operon. In contrast, for functionally unrelated genes, there would be no selection towards coexpression. Therefore, an observation of similar operons found in phylogenetically distant species can be considered an indication of a potential functional relationship between the corresponding genes, even if these genes are scattered in other genomes. Because of the simplicity and elegance of this approach to functional analysis of complete genomes, there are several web sites that offer slightly different approaches to delineation of the conserved gene strings.
WIT/ERGO
The operon comparison tool in the WIT database (http://wit.mcs.anl.gov), the first of the genome context-based tools, was developed by Ross Overbeek in 1998 [640,641]. This tool identifies conserved gene strings by searching for pairs of homologous proteins that are encoded by genes located no more than 300 bp apart on the same DNA strand in each of the analyzed genomes. Each of these pairs is then assigned a score based on the evolutionary distance between the respective species on the rRNA-based phylogenetic tree. It is expected that chance occurrence of pairs of homologous genes in distantly related species is less likely than in closely related ones, so such pairs are more likely to be functionally relevant. Homologous genes are defined as bidirectional best hits in all-against-all BLAST comparisons, which is similar to the method used in constructing the COG database [828].
Because the number of potential gene linkages grows exponentially with the number of the analyzed genomes [640], the sensitivity of methods based on the detection of conserved gene strings can be significantly improved by taking into consideration even unfinished genome sequences. For this reason, WIT and ERGO databases include many incomplete genome sequences from the DOE Joint Genome Institute and other sequencing centers. This approach was used in the successful reconstruction of several known metabolic pathways and led to the correct prediction of candidate genes for some previously uncharacterized metabolic enzymes [82,171,641]. Unfortunately, while this book was in preparation, the ERGO database has been closed for the public, while WIT was still missing some of the useful functionality. We will therefore illustrate the use of the method by exploiting a somewhat similar tool in the COG database.
COGs
The COG database (http://www.ncbi.nlm.nih.gov/COG) allows a simple and straightforward search for conserved operons. Because all proteins in the same COG are presumed to be orthologs, the “Genome context” view, available from each COG page, shows the genes that encode members of the given COG together with the surrounding genes. Genes whose products belong to the same COG are identically colored. This provides for easy identification of sets of COGs that tend to be clustered in genomes. Of course, this tool only works for the genes whose products belong to COGs, so the relationships between genes that are found in only two complete genomes and hence do not belong to any COG would be missed. An exhaustive matching of the co-localization of genes encoding members of the same two COGs allowed new functional predictions for almost 90 COGs, which comprised ~4% of the total set [469,916].
For a practical example of the use of this method, let us consider the search for the archaeal shikimate kinase, the enzyme that is not homologous to the bacterial shikimate kinase (AroK) and hence was not found by traditional sequence similarity searches [171]. Reconstruction of the aromatic amino acids biosynthesis pathway in archaea showed that genomes of A. fulgidus, M. jannaschii, and M. thermoautotrophicum encoded orthologs of bacterial enzymes for all but three reactions of this pathway ([540], see Figure 7.6).
Two of these missing enzymes catalyze first and second reactions of the pathway, indicating that aromatic acids biosynthesis in (most) archaea uses different precursors than in bacteria, whereas the third reaction, phosphorylation of shikimate, was attributed to a non-orthologous kinase, encoded only in archaea [540]. Daugherty and coworkers made a list of the genes involved in aromatic amino acid biosynthesis in archaea and looked for potential neighbors of the aroE gene whose product, shikimate dehydrogenase, catalyzes the reaction immediately preceding the phosphorylation of shikimate (Figure 7.6). In P. abyssi genome, the aroE gene (PAB0300) was followed by an uncharacterized gene (PAB0301) encoding a predicted kinase, which is distantly related to homoserine kinases. This was also the case in A. pernix and T. acidophilum genomes, where the PAB0301-like gene (COG1685, Figure 5.4) was found sandwiched between the aroE gene and the aroA gene, whose product catalyzes the next step of the pathway after shikimate phosphorylation [171]. Genes encoding PAB0301 orthologs (COG1685) were also found in other archaeal genomes, but not in any of the bacterial genomes that contain the typical aroK gene (Figure 5.4). Given this connection, Daugherty et al. expressed MJ1440, the COG1685 member from M. jannaschii and demonstrated that it indeed had shikimate kinase activity [171].
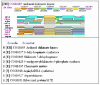
Figure 5.4
Genome context of COG1685 “Archaeal shikimate kinase”. Each line corresponds to an individual genome: aful, Archaeoglobus fulgidus; hbsp, Halobacterium sp.; mjan, Methanococcus jannaschii; mthe, Methanobacterium thermoautotrophicum; pyro, (more...)
STRING
The Search Tool for Recurring Instances of Neighbouring Genes (STRING, http://www.bork.embl-heidelberg.de/STRING), developed by Peer Bork and colleagues, is based on a similar approach [788]. Gene clusters are defined by STRING the same way as in WIT, namely as strings of genes on the same strand located no more than 300 bp from each other. Orthologs are identified as bidirectional best hits using Smith-Waterman comparisons. The STRING search starts from a single protein sequence that can be entered as a FASTA file or just by its gene name in the complete genome. The sequence entered in FASTA format is compared against the database of all proteins encoded in complete genomes so that the user could choose one of the best hits for further examination. Like COGs, STRING contains information only on completely sequenced genomes. The default option in STRING further reduces the number of analyzed genomes by eliminating closely related ones (this option can be switched off by the user). Additionally, STRING features a useful tool that allows the user to perform an “iterative” analysis of gene neighborhoods. After the nearest neighbors of a gene in question are identified, the next “iteration” of STRING would look for their neighbors and record if any of these were found previously. If no new neighbors are found, STRING reports that the search has “converged”. If this does not happen even after five consequent search cycles, the program would just tabulate how many times was each particular gene found in the output. Combined with impressive graphics, this approach makes STRING a fast and convenient tool to search for consistent gene associations in complete genomes.
SNAPper
The SNAP (Similarity-Neighbourhood APproach) tool at MIPS (http://mips.gsf.de/cgi-bin/proj/snap/znapit.pl, [447]) is similar to STRING, but instead of precomputed pairs of orthologs, it simply looks for BLAST hits with user-defined E-values. In addition, SNAP does not require the related genes to form conserved gene strings, they only need to be in the vicinity of each other. SNAPper looks for the homologs of the given protein, than takes neighbors of the corresponding genes, looks for their homologs, and so on [447]. The program then builds a similarity-neighborhood graph (SN-graph), which consists of the chains of orthologous genes in different genomes and adjacent genes in the same genome. The hits that form a closed SN-graph, i.e. recognize the original set of homologs, are predicted to be functionally related. The advanced version of SNAPper offers the choice of several parameters, which allow fine-tuning the performance of the tool depending on the particular query protein.
KEGG
In contrast to the tools described above, identification of gene strings in the KEGG database (http://www.genome.ad.jp/kegg-bin/mk_genome_cmp_html) is geared toward an analysis of the operon conservation. It allows one to find all genes in any two selected complete genomes whose products are sufficiently similar to each other and are separated by no more than five genes. The user can specify the desired degree of similarity between the proteins in terms of the minimal pairwise BLAST score (or maximal Evalue), the minimal length of the alignment, and the type of BLAST hits (bidirectional or unidirectional hits, or just any hits with the specified BLAST score). The user can also specify maximum allowable distances between the genes in either organism, limiting it to any number of genes from zero to five. This option allows one to retrieve much more distant gene pairs than those detected by the ERGO tool. The downside of this richness is that unless one uses fairly strict criteria for protein similarity and the intergenic distances, he or she will end up with dozens or even hundreds of reported gene pairs, few of which would have predictive power. Nonetheless, a sensible use of this tool can bring some very interesting results [268].
Genome context tools in genome annotation
To evaluate the power of gene order-based methods for making functional predictions, we have isolated those cases where a substantial functional prediction did not appear possible without explicit use of gene adjacency information [916]. In spite of the inherent subjectivity of such assessments, the result was instructive: such unique predictions were made for ~90 genes (more precisely, COGs) or ~4% of all COGs analyzed. Given that, as noted above, homology-based approaches already allow functional predictions for a majority of the genes in each sequenced prokaryotic genome, this places gene-string analysis in the position of an important accessory methodology in the hierarchy of genome annotation approaches. Other genome context-based methods may also be useful but are clearly less powerful. This is, of course, a pessimistic assessment because more subtle changes in prediction for gene already annotated by homology-based methods were not taken into account.
These limitations notwithstanding, some of the predictions made on the basis of gene order conservation combined with homology information seem to be exceptionally important. Perhaps the most straightforward case is the prediction of the archaeal exosome, a complex of RNAses, RNA-binding proteins and helicases that mediates processing and 3’->5’ degradation of a variety of RNA species [469]. This finding was made by examination of archaeal genome alignments, which led to the detection of a large superoperon, which, in its complete form, consists of 15 genes. This full complement of co-localized genes, however, is present in only one species, M. thermoautotrophicum, whereas, in all other archaea, the superoperon is partially disrupted and, in some cases, certain genes have been lost altogether. Remarkably, the predicted exosomal superoperon also includes genes for proteasome subunits. According to the logic outlined above, this points to a hitherto unknown functional and possibly even physical association between the proteasome and the exosome, the machines for controlled degradation of RNA and proteins, respectively.
Gene order-based functional prediction seems to be impossible for eukaryotes because of the apparent lack of clustering of functionally linked genes. However, several operons that have been identified in C. elegans [645,894,944] comprise the first exceptions to this rule and suggest that gene order analysis could be eventually used for eukaryotes, too. Besides, the above prediction of proteasome-exosome association might potentially extend to eukaryotes, offering yet another example of the use of prokaryotic genome comparisons for understanding the eukaryotic cell.
Given the fluidity of gene order in prokaryotes, detection of subtle conservation patterns requires fairly sophisticated computational procedures that search for gene neighborhoods , sets of genes that tend to cluster together in multiple genomes, but do not necessarily show extensive conservation of exact gene order [447,491,640,641,709]. One of the interesting findings that have been made possible through these approaches is the prediction of a new DNA repair system in archaeal and bacterial hyperthemophiles [541]. As shown in Figure 5.5 (see color plates), the gene neighborhood predicted to encode this system forms a complex patchwork, with very few conserved gene strings. However, the overall conservation of the neighborhood is obvious (once the analysis is completed and the results are summarized as in Figure 5.5) and statistically significant [541,709]. In an already familiar theme, prediction of this repair system involved a combination of genomic neighborhood detection with fairly complicated protein sequence analysis and structure prediction. One of the notable findings was the identification of a novel family of predicted DNA polymerases (COG1353). Finally, this is where we encounter, once again, COG1518, the protein family already discussed in 4.5. When we first analyzed those proteins, we were inclined to predict that they were novel enzymes, perhaps with a hydrolytic activity. Context analysis allows us to make a much more specific prediction: these proteins mostly likely are nucleases involved in DNA repair.
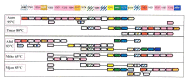
Figure 5.5
Predicted DNA repair system in hyperthermophiles. The pink boxes show optimal growth temperatures for each of the analyzed species (A. aeolicus, T. maritima, A. fulgidus, M. thermoautotrophicum, M. jannaschii). The genes are not drawn to scale; arrows (more...)
5.3. Conclusions and Outlook
In this chapter, we discussed both traditional methods for genome annotation based on homology detection and newer approaches united under the umbrella of genome context analysis. We noted that, although functions can be predicted, at some level of precision, for a substantial majority of genes in each sequenced prokaryotic genome, current annotations are replete with inaccuracies, inconsistencies and incompleteness. This should not be construed as any kind of implicit criticism of those researchers who are involved in genome annotation: the task is objectively hard and is getting progressively more difficult with the growth of databases (and accumulation of inconsistencies). Fortunately, we believe that the remedy is already at hand (see 3.1.3). Specialized databases, designed as genome annotation tools, seem to be capable of dramatically improving the situation, if not solving the annotation problem completely. Prototypes of such databases already exist and function and their extensive growth in the near future seems assured.
The context-based methods of genome annotation are quite new: the development of these approaches started only after multiple genome sequences became available. These approaches have a lot of appeal because they are, indeed, true genomic methods based on the notion that the genome (and, especially, many compared genomes) is much more than the sum of its parts. The results produced by these methods are often very intuitive and even visually appealing as in gene string analysis. Objectively, however, these methods yield considerably less information on gene function than homology-based methods, at least for the foreseeable future. Nevertheless, different genome context approaches substantially complement each other and homology-based methods. In fact, homology-based and context-based methods often produce different and complementary types of functional predictions. The former tend to predict biochemical functions (activities), whereas the latter result in biological predictions, such as involvement of a gene in a particular cellular process (e.g. DNA repair in the example above), even if the exact activity cannot be predicted.
We would like to end this chapter on an upbeat note by stating, in large part on the basis of personal experience, that genome annotation is not a routine, mundane activity as it might seem to an outside observer. On the contrary, this is exciting research, somewhat akin to detective work, which has the potential of teasing out deep mysteries of life from genome sequences.
5.4. Further Reading
- 1.
- Brenner S. Errors in genome annotation. Trends in Genetics. 1999;15:132–133. [PubMed: 10203816]
- 2.
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nature Biotechnology. 2000;18:609–613. [PubMed: 10835597]
- 3.
- Huynen MA, Snel B. Gene and context: integrative approaches to genome analysis. Advances in Protein Chemistry. 2000;54:345–379. [PubMed: 10829232]
- 4.
- Huynen MA, Snel B, Lathe W, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Research. 2000;10:1204–1210. [PMC free article: PMC310926] [PubMed: 10958638]
- 5.
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Research. 2001;11:356–372. [PubMed: 11230160]
- 6.
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Research. 2002;30:482–496. [PMC free article: PMC99818] [PubMed: 11788711]
- 7.
- Ouzounis CA, Karp PD. 2002. The past, present and future of genome-wide re-annotation. Genome Biology 3, COMMENT2001. [PMC free article: PMC139008] [PubMed: 11864365]
Publication Details
Copyright
Publisher
Kluwer Academic, Boston
NLM Citation
Koonin EV, Galperin MY. Sequence - Evolution - Function: Computational Approaches in Comparative Genomics. Boston: Kluwer Academic; 2003. Chapter 5, Genome Annotation and Analysis.




