NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
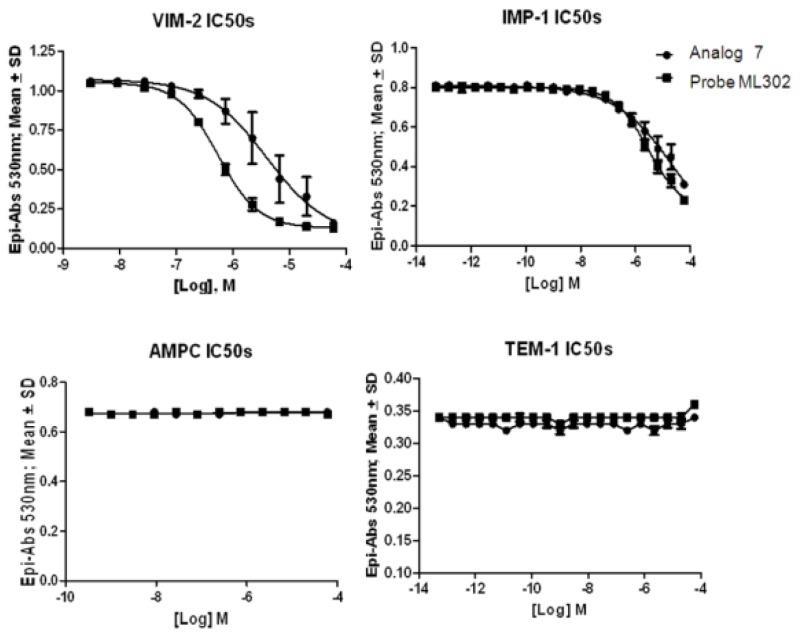
VIM-2 and IMP-1 are Ambler class B metallo-β-lactamases (MBL) capable of hydrolyzing a broad-spectrum of β-lactam antibiotics. Although the discovery and development of MBL inhibitors continue to be an area of active research, an array of potent, non-selective (broad-acting) small molecule inhibitors is yet to be fully characterized. Here we describe a novel dual VIM-2/IMP-1 inhibitor that exhibits efficacy in multiple clinical isolates. We therefore claim CID 53362017/SID 134220672 as a potent, broad acting VIM-2/IMP-1 probe (ML302). This compound was discovered from a medicinal chemistry effort that sought to improve the potency and efficacy of high-throughput screening (HTS) hits. During these chemistry efforts, we identified a rhodanine scaffold that exhibited activity against recombinant VIM-2 and IMP-1 in nitrocefin-based enzyme activity assays. Various secondary assays were run to determine its dual potency (VIM-2 IC50 = 548 nM; IMP-1 IC50 = 3.02 μM) and class B selectivity (it was inactive in TEM-1 and AmpC beta-lactamase enzymatic assays). Kinetic analyses demonstrated that ML302 behaves as a mixed mode uncompetitive/non-competitive inhibitor, with submicromolar Ki values against VIM-2 and IMP-1 (Ki = 183 ± 24 nM and 930 ± 97 nM, respectively). This represents an improvement in both our prior probe ML121 and the previously existing art compound Mitoxantrone, each of which inhibited only VIM-2. Subsequent studies revealed that this probe potentiates the activity of imipenem antibiotic in inhibiting growth of laboratory E. coli BL21 strains harboring VIM-2 and IMP-1, as well as clinical isolates YMC07 (VIM-2-containing Acinetobacter sp.), BAA-2146 (NDM-1-containing Klebsiella pneumonia), PA641 (VIM-2-containing Pseudomonas aeruginosa), and KN20 (IMP-1-containing Pseudomonas aeruginosa). This probe will serve as a valuable tool to elucidate the role of VIM-2 and IMP-1 in nosocomial beta-lactam antibiotic resistance.
Assigned Assay Grant #: 1 R21 NS059451-01 Fast Track
Screening Center Name & PI: Scripps Research Institute Molecular Screening Center (SRIMSC); Hugh Rosen
Chemistry Center Name & PI: SRIMSC; Hugh Rosen
Assay Submitter & Institution: Peter Hodder, TSRI
PubChem Summary Bioassay Identifier (AID): 1854
Probe Structure and Characteristics
| CID/ML# | Target Name | IC50 (nM) [SID, AID] | Anti-target Name | IC50 (μM) [SID, AID] | Fold Selective | Secondary Assays: IC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID 53362017/ML302 | VIM-2 | Enzyme Assays: IC50 = 548 nM [SID 125264855, AID 624079 Ki = 183 nM [SID134220672, AID 624083] | IMP-1 | Enzyme Assays: IC50 = 3.018 μM [SID 125264855, AID 624085] Ki = 930 nM [SID134220672, AID 624084] | >5.5 | Enzyme counterscreens: TEM-1 IC50 >60 μM [SID125264855, AID 624092] AmpC IC50 >60 μM [SID125264855, AID 624090] Imipenem Synergy assays: measured as a reduction (potentiation) of the MIC of imipenem in the following bacterial strains: YMC07/8/B3323 (VIM-2) Achieves 32X Potentiation at 781 nM [SID 125264855, AID 624097] BL21 (VIM-2) Achieves 8X Potentiation at 3.125 μM [SID 125264855, AID 624081] PA641 (VIM-2) Achieves 4X Potentiation at 50 μM [SID 125264855, AID 624080] BAA-2146 (NDM-1) Achieves 2X Potentiation at >50 μM [SID 125264855, AID 624082] KN20 (IMP-1) Achieves 2X Potentiation at >50 μM [SID 125264855, AID 624095] BL21 (IMP-1) Achieves 4X Potentiation at 781 nM [SID 125264855, AID 624097] |
Recommendations for scientific use of the probe
Limitations in state of the art. We previously submitted a probe report describing a selective VIM-2 inhibitor (ML121). This probe was potent and active against the VIM-2 target. However, it lacked activity against the IMP-1 enzyme, which might limit its usefulness in clinically resistant bacterial strains that may express more than one β-lactamase. Another non-competitive VIM-2 inhibitor, mitoxantrone (1,4-dihydroxy-5,8-bis([2-([2-hydroxyethyl]amino)ethyl]amino)-9,10-anthracenedione; CID 4212), is a type II topoisomerase inhibitor that disrupts DNA synthesis and DNA repair in both healthy cells and cancer cells [1]. Like ML121, its inhibition is also selective to VIM-2 and its intense color limits Mitoxantrone’s use in certain assay detection formats. Another prior art compound, 4-chloromercuribenzoic acid (pCMB; CID 1730), is a slowly reversible/irreversible VIM-2 inhibitor shown to have a synergistic effect with β-lactam antibacterials in VIM-2-expressing bacteria.[2] Unfortunately, this cysteine-reactive reagent is known to have several off-target activities, lessening its value for mechanistic studies.
Probe Applications. Here we demonstrate that our probe ML302 (CID 53362017/SID 134220672/SR-03000002555) blocks the enzymatic activities of VIM-2 and IMP-1 in biochemical assays, with no apparent activity against class A (TEM-1) and class C (AmpC) b-lactamases. When dosed in clinical isolates, ML302 shows no toxicity; when co-dosed with imipenem, ML302 significantly increases imipenem’s efficacy (MIC). These findings have significant implications for studies that probe the enzymology of VIM-2 and IMP-1, especially mechanistic studies to better understand these enzymes’ broad-spectrum activity against various beta-lactam based antibiotics. Further, this probe can be useful for experiments that aim to inhibit VIM-2 and IMP-1 activity, without inhibiting TEM-1 or AmpC activity. In a broader role, the non-selectivity, potency, and efficacy of this compound will enable further investigations into the biological and biochemical roles of metallo-β-lactamase enzymes, and may be useful in the design of inhibitors to inhibit these clinically relevant enzymes and reduce the public health burden of antibiotic resistance.
Expected end-users of the probe in the research community. The probe can be used by researchers studying microbiology, antibiotic chemistry, β-lactamase enzymology. Thus, it is conceivable that scientists in diverse fields will be able to apply ML302 to elucidate the role of VIM-2 and IMP-1 in bacterial resistance pathways and investigate mechanisms of VIM-2 and IMP-1 inhibition in biochemical and microbiology-based assays.
Relevant biology of the probe. VIM-2 and IMP-1 are a zinc-dependent, Ambler Class “B” β-lactamases that hydrolyze β-lactam based antibiotics (e.g. penicillins, carbapenems), rendering them ineffective. No VIM-2 inhibitors yet exist for clinical use, and all VIM-2 inhibitors reported to date have been designed to bind zinc or modify cysteine found in the enzyme’s active site. Further, no dual-acting inhibitors exist which block activity of both of these enzymes. Therefore, selective, non-competitive VIM-2 inhibitors are desired to probe VIM-2 function exclusive of active site inhibition.
1. Introduction
The emergence of Gram-negative bacteria that exhibit multi-drug resistance, combined with the lack of new antibiotics, poses a public health challenge [3]. The production of bacterial β-lactamase enzymes, in particular, is a common mechanism of drug resistance [4–6]. The β-lactamases evolved from bacteria with resistance to naturally-occurring β-lactams or penams [7], agents which inhibit the transpeptidase involved in cell wall biosynthesis [8]. Human medicine adapted these agents into synthetic antibiotics such as penicillins, cephalosporins, carbapenems, and monobactams that contain a 2-azetidone ring [7, 9]. The metallo-β-lactamases (MBL) are zinc-dependent class B β-lactamases that hydrolyze the β-lactam ring, rendering the antibiotic ineffective [8, 10]. Increasingly, nosocomial beta-lactam antibiotic resistance arises in P. aeruginosa, Enterobacteriaceae, and other pathogenic bacteria via gene transfer of B1 MBLs [6, 11], including IMP (active on IMiPenem) [12] and VIM (Verona IMipenemase) [13, 14]. For two of these enzymes, VIM-2 and IMP-1, no inhibitors exist for clinical use [8, 11]. Thus, the identification of MBL inhibitors would provide useful tools for reducing nosocomial infections and elucidating their mechanism of action [2, 15]. In the present report, we identify and characterize a compound belonging to a novel rhodanine scaffold that inhibits both VIM-2 and IMP-1 with submicromolar Ki values. This represents a significant advance in the field of broad acting β-lactamase inhibitors.
2. Materials and Methods
2.1. Assays
Table 1Assays from the HTS campaign and prior probe discovery effort (ML121)
| AID | Assay Name | Assay Type | Target | Powder Sample |
|---|---|---|---|---|
| 1527 | Primary biochemical HTS assay to identify inhibitors of VIM-2. | Primary Assay (1X%INH) | VIM-2 | No |
| 1556 | Epi-absorbance primary biochemical HTS assay to identify inhibitors of IMP-1 metallo-beta-lactamase. | Primary Assay (1X%INH) | IMP-1 | No |
| 1856 | Epi-absorbance-based counterscreen for selective VIM-2 inhibitors: biochemical HTS assay to identify inhibitors of IMP-1 metallo-beta-lactamase. | Counterscreen (3X%INH) | IMP-1 | No |
| 1857 | FRET-based counterscreen assay for selective VIM-2 inhibitors: biochemical HTS assay to identify epi-absorbance assay artifacts. | Counterscreen (3X%INH) | VIM-2 (CCF2) | No |
| 1860 | Epi-absorbance-based confirmation biochemical HTS assay to identify selective inhibitors of VIM-2 metallo-beta-lactamase. | Confirmation (3X %INH) | VIM-2 | No |
| 1866 | Epi-absorbance-based counterscreen assay for selective VIM-2 inhibitors: biochemical HTS assay to identify inhibitors of TEM-1 serine-beta-lactamase. | Counterscreen (3X%INH) | TEM-1 | No |
| 1919 | Epi-absorbance-based dose response biochemical high throughput screening assay for selective inhibitors of VIM-2 metallo-beta-lactamase. | Dose Response (3X IC50) | VIM-2 | No |
| 1920 | Epi-absorbance-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of IMP-1 metallo-beta-lactamase. | Dose Response Counterscreen (3X IC50) | IMP-1 | No |
| 1925 | Epi-absorbance-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of TEM-1 serine-beta-lactamase. | Dose Response Counterscreen (3X IC50) | TEM-1 | No |
| 1926 | FRET-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify epi-absorbance assay artifacts. | Dose Response Counterscreen (3X IC50) | VIM-2 (CCF2) | No |
| 1927 | FRET-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of IMP-1 metallo-beta-lactamase. | Dose Response Counterscreen (3X IC50) | IMP-1 (CCF2) | No |
| 1854 | Summary of probe development efforts to identify selective inhibitors of VIM-2 metallo-beta-lactamase. | Summary | VIM-2 | No |
| 2128 | Late stage results from the probe development efforts to identify inhibitors of VIM-2: probe results | Late Stage AID (probe) | VIM-2 | Yes |
| 2317 | Late stage results from the probe development efforts to identify selective inhibitors of VIM-2 metallo-beta-lactamase: Prior art results. | Late Stage AID (prior art) | VIM-2 | Yes |
| 504620 | Late stage Assay provider assay to determine imipenem Synergy of probe ML121 | Late Stage AID (probe ML121) | VIM-2 | Yes |
Table 2Assays for the current probe discovery effort (ML302)
| AID | Assay Name | Assay Type | Target | Powder Sample |
|---|---|---|---|---|
| 624079 | Absorbance-based Biochemical Nitrocefin Substrate Hydrolysis Assays (IC50) | Dose Response (2X IC50) (probe ML302) | VIM-2 | Yes |
| 624085 | IMP-1 | Yes | ||
| 624092 | TEM-1 | Yes | ||
| 624090 | AmpC | Yes | ||
| 624083 | Absorbance-based Biochemical Nitrocefin Substrate Hydrolysis Assays (Ki and Km) | Dose Response (2X) | VIM-2 | Yes |
| 624084 | IMP-1 | Yes | ||
| 624096 | Absorbance based Imipenem Antibiotic Synergy Assay (MIC4X): YMC07/B3323 | Dose Response (2X) | VIM-2 | Yes |
| 624081 | Absorbance based Imipenem Antibiotic Synergy Assay (MIC4X): BL21 | Dose Response (2X) | VIM-2 | Yes |
| 624080 | Absorbance based Imipenem Antibiotic Synergy Assay (MIC4X): PA641 | Dose Response (2X) | VIM-2 | Yes |
| 624082 | Absorbance based Imipenem Antibiotic Synergy Assay (MIC4X): BAA-2146 | Dose Response (2X) | NDM-1 | Yes |
| 624095 | Absorbance based Imipenem Antibiotic Synergy Assay (MIC4X): KN20 | Dose Response (2X) | IMP-1 | Yes |
| 624097 | Absorbance based Imipenem Antibiotic Synergy Assay (MIC4X): BL21 | Dose Response (2X) | IMP-1 | Yes |
(also see Summary AID 1854)
VIM-2 Inhibition Assays (Epi-Absorbance-based; Nitrocefin) (PubChem AIDs: AID 1527,AID 1860, AID 1919, AID 2128, AID 2317, and AID 624079). The purpose of this assay is to identify compounds that act as inhibitors of the VIM-2 β-lactamase. This biochemical epi-absorbance-format assay employs the cephalosporin nitrocefin as the VIM-2 substrate, and takes advantage of the fluorescent properties of white microtiter plates [16]. Nitrocefin is a yellow chromogenic substrate (Imax = 395 nm) that is hydrolyzed by β-lactamases to yield a red product with increased absorbance properties (Imax = 495 nm) that quenches plate fluorescence by absorbing the plate’s emission light [16]. In this assay, test compounds are incubated with purified VIM-2 enzyme and nitrocefin in detergent-containing buffer at room temperature. The reaction is stopped by the addition of EDTA, followed by measurement of well fluorescence. As designed, compounds that inhibit VIM-2 will inhibit nitrocefin hydrolysis, inhibit generation of red product, and inhibit quenching of plate fluorescence, resulting in an increase in well fluorescence. Compounds were tested in singlicate (AID 1527) and triplicate (AID 1860) at a final nominal concentration of 5.6 μM, in a 10-point 1:3 dilution series starting at a nominal concentration of 55.7 μM (AID 1919, AID 2128, AID 2317, and AID 624079).
IMP-1 Inhibition Counterscreens (Epi-absorbance-based; Nitrocefin) (AID 1556, AID 1856, AID 1920, AID 2128, AID 2317, and AID 624085). The purpose of this assay is to identify compounds that act as inhibitors of the IMP-1 β-lactamase. This biochemical epi-absorbance-format assay employs the cephalosporin nitrocefin as the IMP-1 substrate, and takes advantage of the fluorescent properties of white microtiter plates [16]. This assay also serves as a counterscreen to determine whether compounds identified as possible VIM-2 selective inhibitors are non-selective due to inhibition of IMP-1. Nitrocefin is a yellow chromogenic substrate (Imax = 395 nm) that is hydrolyzed by β-lactamases to yield a red product with increased absorbance properties (Imax = 495 nm) that quenches plate fluorescence by absorbing the plate’s emission light [16]. In this assay, test compounds are incubated with purified IMP-1 enzyme and nitrocefin in detergent-containing buffer at room temperature. The reaction is stopped by the addition of EDTA, followed by measurement of well fluorescence. As designed, compounds that inhibit IMP-1 will inhibit nitrocefin hydrolysis, inhibit generation of red product, and inhibit quenching of plate fluorescence, resulting in an increase in well fluorescence. Compounds were tested in singlicate (AID 1556) or in triplicate (AID 1856) at a final nominal concentration of 5.6 μM, and in triplicate using a dilution series starting at a nominal test concentration of 60 micromolar (AID 1920, AID 2128, AID 2317, and AID 624085).
2.2. Probe Chemical Characterization
The synthesis of the probe and analytical characterization data are probe ML302 are provided in Section 2.3. ML302 was obtained with >98% purity according to 1H NMR and LCMS analysis. However, ML302 had relatively poor solubility characteristics, so we also generated a more soluble HCl salt, identified as CID53384679. The HCl salt was also obtained with >96% purity by 1H NMR and LCMS analysis.
Solubility. The solubility of probe ML302 (synthesized and registered by the SRISMC as SR-03000002555-2/SID 134220672/CID 53362017) was measured in phosphate buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic and a pH of 7.4) at room temperature (23°C). The solubility of was found to be <1 μM. In an attempt to achieve improved solubility, a series of salts (including the HCl salt, CID 53384679) were synthesized and studied. However, salt HCl CID 53384679 also had poor solubility (<1 μM) in this standard pH 7.4 PBS buffer assay system. However, probe ML302 and HCl salt CID53384679 are fully soluble at 100 μM in 70:30 DMSO/water, and both have solubility >100 μM at pH = 3 aqueous solution. A sample of the HCl salt CID 53384679 is also soluble >100 μM in D2O (1H NMR measurements performed at this concentration). Both probe ML302 and the HCl salt CID 53384679 are fully soluble under the conditions of the VIM-2 biochemical assays and antibacterial assays described in this probe report.
Stability. The stability of probe ML302 was measured at room temperature (23ºC) in PBS (no antioxidants or other protectants; DMSO concentration below 0.1%). The stability, represented by the half-life, was found to be very poor. Figure 1 provides graphs showing loss of compound with time over a 48 hour period with a minimum of 6 time points. The table at the end of this section indicates the percent of compound remaining at the end of the 48 hours. These data suggest that the probe ML302 and HCl salt CID 53384679 are highly unstable under these assay conditions. However, these data are not consistent with our experience with these compounds when handled under other conditions. Rather, we suspect that these data reflect solubility problems with the compound under the assay conditions. For example, HCl salt CID 53384679 proved to be fully stable, at 100 μM, in D2O over a two-week monitoring period (1H NMR study)—conditions under which it is fully soluble. Similarly, probe ML302 and HCl salt CID 53384679 were fully stable at 100 μM in 70/30 DMSO-D2O over a two-week period, in presence of air (1H NMR study)—conditions, again, under which they are fully soluble. ML302 and HCl salt CID 53384679 (both 1 μM) were fully stable (24 h monitoring) at pH 3 in PBS with 20% DMSO, and displayed >93% stability (over 24 h) at pH 7 PBS with 20% DMSO. The latter value undoubtedly reflects the limit of solubility of these compounds under these conditions. Apparently, the HCl salt CID 53384679 is converted to the free base (ML302) at pH 7.4.
Probe ML302, and its HCl salt were measured for its ability to form glutathione adducts. At concentrations of 100 μM reduced GSH, 10 μM of the new probe does not appear to be a Michael acceptor [17, 18]. Evidently, the chlorine substituents at the 2,6-position of the phenyl ring hinder the double bond such that these compounds are not highly active as Michael acceptors.
Table 3Solubility of ML302
| Compound | MW | SR Number | CID | SID | Solubility in PBS (μM) | Michael Acceptor 100 μM GSH trap | Stability in PBS t1/2 (hr) |
|---|---|---|---|---|---|---|---|
| New Probe ML302 | 479.8 | SR-03000002555-2 | 53362017 | 134220672 | <1 μM | No | Section 2.2 |
| Probe ML302 HCl salt | 516.3 | SR-03000002594-2 | 53384679 | 134220676 (synthesis) | <1 μM | No | Section 2.2 |
2.3. Probe Preparation
VIM-2 inhibitor probe ML302 was synthesized by a two-step procedure involving the condensation of 2,3,6-trichlorobenzaldehyde with rhodanine-3-acetic acid, followed by coupling of the product carboxylic acid 1 with 1-amino-4-methyl-piperazine [19].
2,3,6-Trichlorobenzaldehdye (1.676 g, 8.0 mmol, 1 equiv, purchased from Oakwood Product, Inc. # 043332) and rhodanine-3-acetic acid (1.530 g, 8.0 mmol, 1 equiv, purchased from Alfa Aesar, # B22244-06) were weighed into a 20-mL microwave vial. Ethanol (16 mL, to give a reaction concentration of 0.5 M) was added followed by piperidine (4 drops). The vial was sealed and submitted to microwave irradiation at 120 °C for 1 h. The solvent was then removed and the crude product was recrystallized from hot ethanol and water. The solid was filtered, rinsed with water and dried under high vacuum yielding compound 1 as a yellow solid (2.59 g, 85%): 1H NMR (400 MHz, d6-DMSO) δ 13.56 (1H, br s), 7.84 (1H, s), 7.87-7.84 (1H, m), 7.81 (1H, d, J = 8.8 Hz), 7.67 (1H, d, J = 8.7Hz), 4.70 (2H, s). 13C NMR (100 MHz, d6-DMSO) δ 192.6, 167.2, 164.9, 133.0, 132.2, 131.5, 131.4, 131.2, 131.1, 129.8, 128.6, 45.6.
A solution of 1 (76 mg, 0.2 mmol, 1 equiv.) and BOPCl (61 mg, 0.24 mmol, 1.2 equiv) in dichloromethane (2 mL, to give a reaction concentration of 0.1 M) in a 5-mL round-bottomed flask was cooled to 0 °C using an ice/water bath under an inert atmosphere. Freshly distilled triethylamine (35 μL, 0.24 mmol, 1.2 equiv) was added and the solution was allowed to stir at 0 °C for 30 min. The cold bath was removed and 1-amino-4-methyl-piperazine (29 μL, 0.24 mmol, 1.2 equiv, purchased from Acros Organics, # 251391000) was added. The mixture was allowed to warm to room temperature and was stirred overnight. The crude product, obtained by removal of all solvents volatile compounds in vacuo, was directly purified by flash chromatography (silica gel, dichloromethane-methanol 90/10, Rf = 0.29) yielding compound 2 as an inseparable mixture of E/Z isomers in a 1:3 ratio (75 mg, 78 %, yellow solid, mp = 184–186 °C) (Figure 2). The chemical purity of 2 is >98% according to HLPC analysis.
Data for VIM-2 Inhibitor Probe, CID 53362017 (Compound 2), Z isomer (major): 1H NMR (400 MHz, d6-DMSO) δ 9.08 (1H, s), 7.83 (1H, s), 7.82 (1H, d, J = 8.7 Hz), 7.68 (1H, d, J = 8.8 Hz), 4.91 (2H, s), 2.99-2.62 (6H, m), 2.43-2.02 (2H, m), 2.17 (3H, s); 13C NMR (100 MHz, d6-DMSO) δ 192.8, 166.0, 165.1, 133.0, 132.2, 131.5, 131.4, 131.3, 131.2, 129.8, 128.2, 55.1, 54.3, 45.3, 45.2. MS ([M+H]+): 481.1 (100%), 479.4 (71%), 483.0 (39%). IR (cm−1): 3049, 2927, 2808, 1731, 1687, 1627, 1402, 1366, 1323, 1286, 1273, 1193, 1175, 1051, 1008, 816, 725.
Partial Data for (minor) E isomer of VIM-2 Inhibitor Probe CID 53362017: 1H NMR (400 MHz, d6-DMSO) δ 9.37 (1H, s), 7.82 (1H, d, J = 8.7 Hz), 7.81 (1H, s), 7.68 (1H, d, J = 8.8 Hz), 4.56 (2H, s), 2.99-2.62 (6H, m), 2.43-2.02 (2H, m), 2.15 (3H, s).
All anti-bacterial and enzyme assays performed with the VIM-2 inhibitor probe, CID 53362017 (compound 2) were performed with the 3:1 mixture of olefin isomers. The two olefin isomers of the Probe were not separated, since prior studies with three probe analogs (SR-6818, SR-2448 and SR-2450) (Figure 3) indicated that the separated olefin isomers rapidly re-isomerized to the original 3:1 mixtures. (The olefin isomers of SR-6818, SR-2448 and SR-2450 were separated by preparative HPLC. In all three cases, the separated isomers had converted back to the original 3:1 mixtures in the time required to concentrate the HPLC fractions prior to NMR analysis).
A solution of hydrogen chloride in dioxane (4.0M, 4.1 mL, 16.4 mmol, 80 equiv) was slowly added at 0 °C to compound 2 (100 mg, 0.21 mmol, 1 equiv) under an inert atmosphere. The cold bath was removed and the mixture was allowed to stir at room temperature for 4 h. The resulting heterogeneous solution was filtered and the yellow precipitate 3 (Figure 4) was thoroughly washed with diethyl ether and dried under vacuum. No further purification was performed and the hydrochloride salt was obtained as a mixture of E/Z isomers in a 1:1 ratio (96 mg, 89 %, yellow solid, mp = 203–206 °C).
Data for VIM-2 Inhibitor CID 53384679 (Compound 3), Z isomer: 1H NMR (400 MHz, d6-DMSO) δ 11.21 (1H, br s), 9.41 (1H, s), 7.82 (1H, s), 7.81 (1H, d, J = 8.5 Hz), 7.67 (1H, d, J = 8.5 Hz), 4.99 (2H, s), 3.49-3.00 (8H, m), 2.73 (3H, s); 13C NMR (100 MHz, d6-DMSO) δ 192.8, 165.1, 162.1, 132.9, 132.2, 131.5, 131.4, 131.2, 131.1, 129.8, 128.3, 52.5, 51.8, 50.7, 45.3. MS ([M+H]+): 481.1 (100%), 479.5 (74%), 483.0 (38%). IR (cm−1): 2936, 2464, 1724, 1692, 1564, 1437, 1335, 1190, 1176, 1100, 1055, 984, 898, 814.
Partial Data for E isomer of VIM-2 Inhibitor Compound 3 CID 53384679: 1H NMR (400 MHz, d6-DMSO) δ 11.21 (1H, br s), 9.97 (1H, s), 7.81 (1H, d, J = 8.5 Hz), 7.80 (1H, s), 7.67 (1H, d, J = 8.5 Hz), 4.60 (2H, s), 3.49-3.00 (8H, m), 2.71 (3H, s).
3. Results
3.1. Summary of Screening Results
During the medicinal chemistry effort following our initial VIM-2 selective inhibitor project, the rhodanine scaffold, exemplified by SR-01000216818/CID1733268, exhibited activity against the anti-target enzyme, IMP-1. This suggested that this scaffold could be a source of broader-acting dual inhibitors of bacterial MBLs which might serve as tools for novel antibiotic discovery. In order to assess the efficacy of the new scaffold, we tested powder samples of leads in a series of biochemical and bacterial-based assays. Initial studies with recombinant VIM-2 protein revealed that several compounds in this scaffold exhibited potencies less than 5 micromolar. The broad acting MBL probe (ML302) was identified from this scaffold.
3.2. Dose Response Curves for Probes
Dose response curves for probe ML302 are shown in Figure 5 against all enzymes tested. The probe shows selective potency to the VIM-2 and IMP-1 enzymes. For comparison, SR-01000216818/CID 1733268 (“Analog 7”) is also graphed. Potency values are reported in the cover page of this report.
3.3. Scaffold/Moiety Chemical Liabilities
There is no known instability or chemical liabilities associated with the chemical scaffold of probe ML302. The compounds appear to be fully stable during all routine handling operations (synthesis chromatographic purification of the probe, NMR studies, storage as powders, etc.). Issues with solubility of the probe and its HCl salt are discussed in a previous section. The only issue we have observed is that the probe and its HCl salt are unstable at pH 10, due to amide hydrolysis at this basic pH. Therefore, handling of probe ML302 at pH > 7.4 is not recommended.
3.4. SAR Tables
Describe SAR & chemistry strategy (including structure and data) that led to the probe. Following submission of our initial probe report describing a selective VIM-2 inhibitor probe (ML121), compounds in the rhodanine scaffold appeared to have activity for both VIM-2 and IMP-1, suggesting a broad-acting MBL inhibitor probe could be developed. Powder and re-synthesized samples of selected compounds were tested by the assay provider in several biochemical and bacterial assays to determine potency, efficacy, and selectivity. These efforts resulted in probe ML302 and related analogs. The synthesis of ML302, presented in Section 2.3 of this probe report, is amenable to synthesis of additional analogs by substituting different aldehydes instead of 2,3,6-trichlorobenzaldehyde using the condensation of rhodanine-3-acetic acid; use of different amines instead of 1-amino-4-methyl-piperazine in the coupling with carboxylic acid 1. As summarized in the following SAR tables, more than 90 such analogs have been synthesized and tested. Preliminary efforts indicate that introduction of amino acid side chains in the rhodanine scaffold also lead to active compounds. Additional studies of such analogs will be performed in the future.
Metallo-beta Lactamase Inhibitor SAR Table: Rhodanine Scaffold
| Compound Information | Bioassays | Biochemical Assays | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM- | BAA-2146 (NDM-1) | PA641 (VIM-2) | KN20 (IMP-1) | BL21 (IMP-1) | VIM-2 | IMP-1 | TEM-1 | AmpC | VIM-2 Ki | IMP-1 Ki Assay | ||||||||
| MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | IC50 (uM) | IC50 (uM) | IC50 (uM) | IC50 (uM) | Ki (nM) | Km (uM) | Ki (nM) | Km (uM) | ||||||||
| Probe | SR-03000002555-1 |
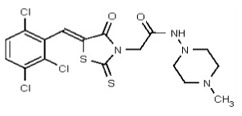 | 53362017 | 125264855 | TSRI | None | 3.125 | 8X | 0.781 | 32X | >50 | 2X | 50 | 4X | >50 | 2X | 0.781 | 0.5481 | 3.018 | >60 | >60 | ND | ND | ND | ND | ||
| Probe ML302 (Sample to NIH) | SR-03000002555-2 |
 | 53362017 | 134220672 | 003940491 | TSRI | None | ND | 0.391 | 32X | ND | ND | ND | ND | ND | ND | ND | ND | 183 +/− 24 | 14.8 +/− 3 Mixed mode inhibitor | 930 +/− 97 | 6.4 +/− 1.3 Mixed mode inhibitor | |||||
| Analog 1 | SR-03000002674-1 |
 | 53495083 | 126723255 | 003940492 | TSRI | None | 1.563 | 0.781 | 16X | >50 | >50 | >50 | 3.125 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 2 | SR-03000002704-1 |
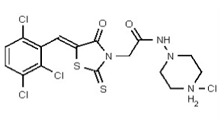 | 54579799 | 131269026 | 003940493 | TSRI | None | 12.5 | 0.781 | 32X | >50 | 2X | >50 | 2X | >50 | 25 | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Analog 3 | SR-03000002705-1 |
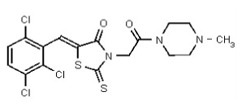 | 54579797 | 131269027 | TSRI | None | 6.25 | 1.563 | 16X | >50 | 50 | 4X | >50 | 0.781 | 8X | ND | ND | ND | ND | ND | ND | ND | ND | ||||
| Analog 3 | SR-03000002705-2 |
 | 54579797 | 134220673 | 003940494 | TSRI | None | ND | 0.781 | 32X | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 4 | SR-03000002672-1 |
 | 53495082 | 126723253 | TSRI | None | 3.125 | 1.563 | 16X | >50 | >50 | >50 | 0.781 | ND | ND | ND | ND | ND | ND | ND | ND | ||||||
| Analog 4 | SR-03000002672-2 |
 | 53495082 | 134220674 | 003940495 | TSRI | None | ND | 1.563 | 32X | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 5 | SR-03000002594-1 |
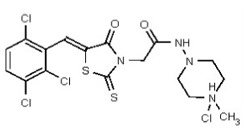 | 53384679 | 125311278 | TSRI | None | 3.125 | 8X | 1.563 | 16X | >50 | >50 | Achievs 2X | >50 | 1.563 | ND | ND | ND | ND | ND | ND | ND | ND | ||||
| Analog 5 | SR-03000002594-2 |
 | 53384679 | 134220676 | 003940496 | TSRI | None | ND | 0.781 | 16X | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Compound Information | Bioassays | Biochemical Assays | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | BAA-2146 (NDM-1) | PA641 (VIM-2) | KN20 | BL21 | VIM-2 | IMP-1 | VIM-2 Ki | ||||
| MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | MIC 4× (μM) | IC50 (uM) | IC50 (uM) | Ki (nM) | ||||||||
| Analog 6 | SR-03000002441-1 |
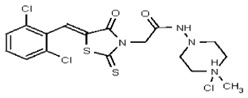 | 53308605 | 124767346 | TSRI | None | 12.5 | 8X | 3.125 | 32X | >100 | 2X | >100 | 2X | >100 | 3.125 | ND | ND | ND | |
| Analog 6 (resynthesized for submission to NIH) | SR-03000002441-2 |
 | 53308605 | 134220675 | 003940497 | TSRI | None | ND | 3.125 | 32X | ND | ND | ND | ND | ND | ND | ND | |||
| Analog 7 (Purchased) | SR-01000216818-4 |
 | 1733268 | 135631809 | ChemBridge | 5680564 | 4.55 | 1.781 +/− 0.384 | ||||||||||||
| Analog 7 (resynthesized) | SR-01000216818-6 |
 | 1733268 | 123083180 | TSRI | None | ND | 25 | Prepared in 75/25 DMSO/H2O | ND | >100 | ND | ND | ND | ND | ND | ||||
| Analog 7 (purchased for reference) | SR-01000216818-7 |
 | 1733268 | 124360376 | ChemBridge | 5680564 | ND | 3.125 | Prepared in neat DMSO | ND | ND | ND | ND | ND | ND | ND | ||||
| Analog 7 (resynthesized) | SR-01000216818-8 |
 | 1733268 | 124384965 | 003940498 | TSRI | None | 6.25 | 8X | 6.25 | 32X | >100 | >100 | 2X | >100 | 25 | 3.98 | 31.9 | ND | |
| Analog 8 | SR-03000002375-1 |
 | 53257035 | 124398381 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 9 | SR-03000002376-1 |
 | 53257030 | 124398382 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 10 | SR-03000002377-1 |
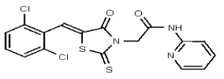 | 53257026 | 124398383 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Compound Information | Bioassays | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | BAA-2146 (NDM-1) | PA641 (VIM-2) | KN20 | BL21 | ||||
| MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | MIC 4× (μM) | ||||||||
| Analog 11 | SR-03000002380-1 |
 | 53257031 | 124398386 | TSRI | None | >100 | ND | ND | ND | ND | ND | |||||
| Analog 12 | SR-03000002381-1 |
 | 53257037 | 124398404 | TSRI | None | 12.5 | 8X | ND | ND | ND | ND | ND | ||||
| Analog 13 | SR-03000002461-1 |
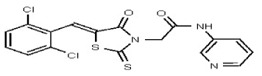 | 1382782 | 125001880 | Vitas-M Lab | STL035211 | ND | 6.25 | 16X | ND | ND | ND | ND | ||||
| Analog 14 | SR-03000002378-1 |
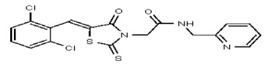 | 53257011 | 124398384 | TSRI | None | 6.2 | 8X | 6.25 | 16X | >100 | >100 | 2X | >100 | ND | ||
| Analog 15 | SR-03000002379-1 |
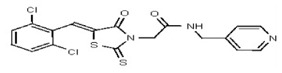 | 53257036 | 124398385 | TSRI | None | 12.5 | 6.25 | 16X | ND | ND | ND | ND | ||||
| Analog 16 | SR-03000002395-1 |
 | 53301882 | 124756530 | TSRI | None | >100 | ND | ND | ND | ND | ND | |||||
| Analog 17 | SR-03000002396-1 |
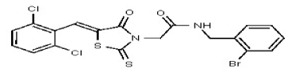 | 53301881 | 124756531 | TSRI | None | >100 | ND | ND | ND | ND | ND | |||||
| Analog 18 | SR-03000002433-1 |
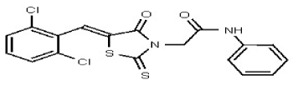 | 53308614 | 124767337 | TSRI | None | >100 | ND | ND | ND | ND | ND | |||||
| Analog 19 | SR-03000002434-1 |
 | 53308615 | 124767338 | TSRI | None | >100 | ND | ND | ND | ND | ND | |||||
| Compound Information | Bacterial Assays (Strain and MBL) | Biochemical Assays | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | BAA-2146 (NDM-1) | PA641 (VIM-2) | KN20 | BL21 | VIM-2 | IMP-1 | VIM-2 Ki | ||||
| MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | MIC 4× (μM) | IC50 (uM) | IC50 (uM) | Ki (nM) | ||||||||
| Analog 20 | SR-03000002455-1 |
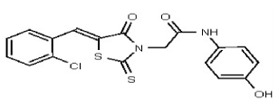 | 2049116 | 125001873 | Innovapharm Ltd. | STT-00175051 | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 21 | SR-03000002460-1 |
 | 5724963 | 125001879 | Vitas-M Lab | STL035210 | 3.13 | 50 | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 22 | SR-03000002385-1 |
 | 53301880 | 124756519 | TSRI | None | 25 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 23 | SR-03000002386-1 |
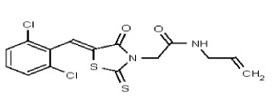 | 53301896 | 124756520 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 24 | SR-03000002398-1 |
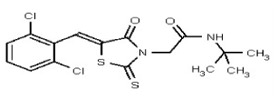 | 53301889 | 124756533 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 25 | SR-03000002447-1 |
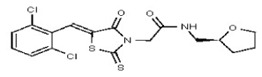 | 53313357 | 124807342 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 26 | SR-03000002448-1 |
 | 53313358 | 124807343 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 27 | SR-03000002457-1 |
 | 17592954 | 125001875 | Vitas-M Lab | STK626562 | 100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 28 | SR-03000002459-1 |
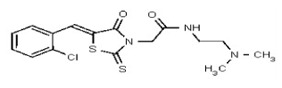 | 50739720 | 125001877 | Innovapharm Ltd. | STT-00362432 | 100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Analog 29 | SR-00000009750-1 |
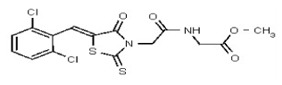 | 53308608 | 124767339 | TSRI | None | >100 | ND | ND | ND | ND | ND | ND | ND | ND | |||||
| Compound Information | Bacterial Assays (Strain and MBL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | ||
| MIC 4× (μM) | Imipenem Potentiation | MIC 4× (μM) | Imipenem Potentiation | ||||||||
| Analog 30 | SR-03000002382-1 |
 | 53257017 | 124398405 | TSRI | None | 12.5 | 8X | 6.25 | 16X | |
| Analog 31 | SR-03000002383-1 |
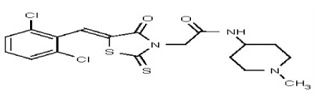 | 53301893 | 124756517 | TSRI | None | 12.5 | 8X | ND | ||
| Analog 32 | SR-03000002432-1 |
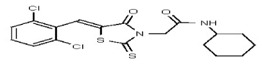 | 53308616 | 124767336 | TSRI | None | >100 | ND | |||
| Analog 33 | SR-03000002374-1 |
 | 1560458 | 124384967 | TSRI | None | 12.5 | 8X | ND | ||
| Analog 34 | SR-03000002431-1 |
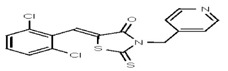 | 53308606 | 124767335 | TSRI | None | 12.5 | 8X | ND | ||
| Analog 35 | SR-03000002442-1 |
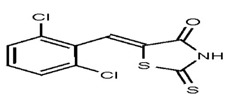 | 1280940 | 124767347 | TSRI | None | 100 | ND | |||
| Analog 36 | SR-03000002458-1 |
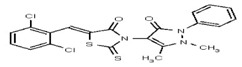 | 1908865 | 125001876 | Vitas-M Lab | STK861771 | 12.5 | 8X | ND | ||
| Analog 37 | SR-03000002397-1 |
 | 53301874 | 124756532 | TSRI | None | >100 | ND | |||
| Analog 38 | SR-03000002454-1 |
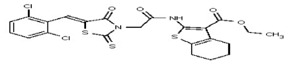 | 1554607 | 125001872 | Innovapharm Ltd. | STT-00172281 | >100 | ND | |||
| Analog 39 | SR-03000002456-1 |
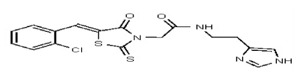 | 6226188 | 125001874 | Vitas-M Lab | STK601034 | >100 | ND | |||
| Compound Information | Bacterial Assays (Strain and MBL) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | BAA-2146 (NDM-1) | PA641 (VIM-2) | KN20 | ||||
| MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | ||||||||
| Analog 40 | SR-03000002462-1 |
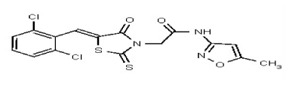 | 5858470 | 125001881 | Vitas-M Lab | STL035212 | 100 | ND | ND | ND | ND | |||||
| Analog 41 | SR-03000002446-1 |
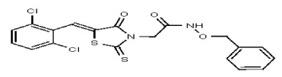 | 53313356 | 124807341 | TSRI | None | >100 | ND | ND | ND | ND | |||||
| Analog 42 | SR-03000002451-1 |
 | 53313350 | 124807346 | TSRI | None | >100 | ND | ND | ND | ND | |||||
| Analog 43 | SR-03000002452-1 |
 | 53338906 | 125001870 | TSRI | None | 6.2 | 8X | 12.5 | 8X | >100 | >100 | >100 | |||
| Analog 44 | SR-03000002449-1 |
 | 53313352 | 124807344 | TSRI | None | >100 | ND | ND | ND | ND | |||||
| Analog 45 | SR-03000002450-1 |
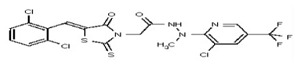 | 53313351 | 124807345 | TSRI | None | >100 | ND | ND | ND | ND | |||||
| Analog 46 | SR-03000002480-1 |
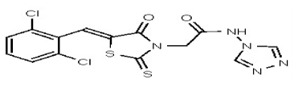 | 53346521 | 125011825 | TSRI | None | 100 | ND | ND | ND | ND | |||||
| Analog 47 | SR-03000002481-1 |
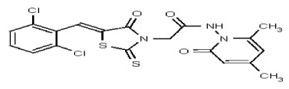 | 53346520 | 125011826 | TSRI | None | ND | 1.563 | 32X | ND | ND | ND | ||||
| Analog 48 | SR-03000002384-1 |
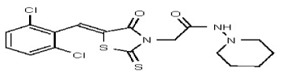 | 53301887 | 124756518 | TSRI | None | 6.2 | 8X | 3.125 | 16X | ND | >100 | >100 | |||
| Analog 49 | SR-03000002453-1 |
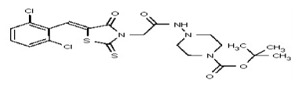 | 53338912 | 125001871 | TSRI | None | 6.2 | 8X | 3.125 | 16X | >100 | >100 | >100 | |||
| Analog 50 | SR-03000002482-1 |
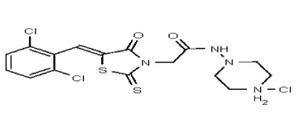 | 53346529 | 125011827 | TSRI | None | 3.125 | 8X | 3.125 | 16X | >50 | >50 | 2X | >50 | ||
| Analog 51 | SR-03000002588-1 |
 | 53384710 | 125311272 | TSRI | None | 12.5 | 16X | 6.25 | 32X | >50 | ND | >50 | |||
| Analog 52 | SR-03000002589-1 |
 | 53384681 | 125311273 | TSRI | None | ND | 3.125 | >50 | >50 | >50 | |||||
| Analog 53 | SR-03000002590-1 |
 | 53384726 | 125311274 | TSRI | None | ND | 6.25 | 32X | >50 | >50 | 2X | >50 | |||
| Analog 54 | SR-03000002591-1 |
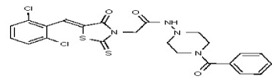 | 53384676 | 125311275 | TSRI | None | ND | 3.125 | 16X | >50 | >50 | >50 | ||||
| Analog 55 | SR-03000002592-1 |
 | 53384686 | 125311276 | TSRI | None | ND | 25 | ND | ND | ND | |||||
| Analog 56 | SR-03000002593-1 |
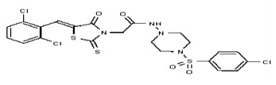 | 53384728 | 125311277 | TSRI | None | ND | >50 | 2X | ND | ND | ND | ||||
| Analog 57 | SR-03000002737-1 |
 | 54669692 | 131465571 | TSRI | None | ND | 25 | 8X | ND | ND | ND | ||||
| Analog 58 | SR-03000002373-1 |
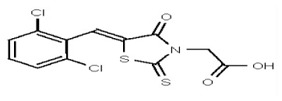 | 1201205 | 124384966 | TSRI | None | 3.125 | 8X | 12.5 | 16X | >100 | >100 | >100 | |||
| Analog 59 | SR-03000002519-1 |
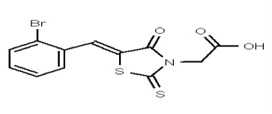 | 1205102 | 125258648 | TSRI | None | ND | >50 | 2X | ND | ND | ND | ||||
| Compound Information | Bacterial Assays (Strain and MBL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | ||
| MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | ||||||||
| Analog 60 | SR-03000002520-1 |
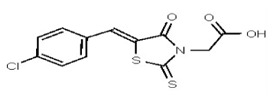 | 1201114 | 125258649 | TSRI | None | ND | >50 | 2X | ||
| Analog 61 | SR-03000002521-1 |
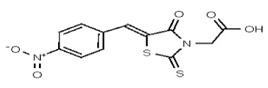 | 1622325 | 125258651 | TSRI | None | ND | >50 | 2X | ||
| Analog 62 | SR-03000002522-1 |
 | 13337884 | 125258652 | TSRI | None | ND | >50 | 2X | ||
| Analog 63 | SR-03000002523-1 |
 | 1201949 | 125258653 | TSRI | None | ND | >50 | |||
| Analog 64 | SR-03000002524-1 |
 | 1201311 | 125258654 | TSRI | None | ND | >50 | |||
| Analog 65 | SR-03000002525-1 |
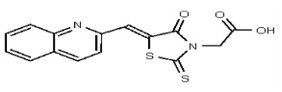 | 5943467 | 125258655 | TSRI | None | ND | >50 | |||
| Analog 66 | SR-03000002526-1 |
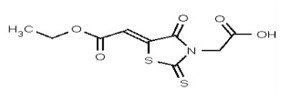 | 53356622 | 125258656 | TSRI | None | ND | >50 | |||
| Analog 67 | SR-03000002527-1 |
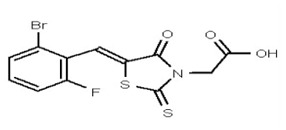 | 53356627 | 125258657 | TSRI | None | ND | >50 | 2X | ||
| Analog 68 | SR-03000002528-1 |
 | 53356652 | 125258658 | TSRI | None | ND | >50 | 2X | ||
| Analog 69 | SR-03000002529-1 |
 | 53356632 | 125258660 | TSRI | None | ND | 6.25 | 16X | ||
| Compound Information | Bacterial Assays (Strain and MBL) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | SR-Number | Structure | CID | SID | MLS | Vendor | Vendor Catalog ID | BL21 (VIM-2) | YMC07/B3323 (VIM-2) | BAA-2146 (NDM-1) | PA641 (VIM-2) | KN20 | ||||
| MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | Imipenem Potentiation | MIC 4x (μM) | ||||||||
| Analog 70 | SR-03000002530-1 |
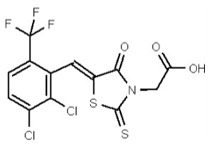 | 53356630 | 125258661 | TSRI | None | ND | 6.25 | 16X | ND | ND | ND | ||||
| Analog 71 | SR-03000002531-1 |
 | 53356624 | 125258662 | TSRI | None | ND | 6.25 | 8X | ND | ND | ND | ||||
| Analog 72 | SR-03000002586-1 |
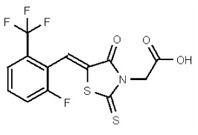 | 53384688 | 125311270 | TSRI | None | 25 | 8X | 25 | 2x | >50 | ND | >50 | |||
| Analog 73 | SR-01000199849-2 |
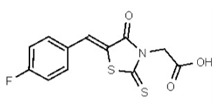 | 1381767 | 125258650 | TSRI | None | ND | >50 | ND | ND | ND | |||||
| Analog 74 | SR-01000214156-2 |
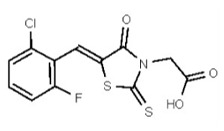 | 1201186 | 125258659 | TSRI | None | ND | >50 | 2X | ND | ND | ND | ||||
| Analog 75 | SR-01000686826-3 |
 | 1594367 | 125001878 | Vitas-M Lab | STK994030 | 50 | ND | ND | ND | ND | |||||
| Analog 76 | SR-01000699784-2 |
 | 1553748 | 125001882 | Vitas-M Lab | STL035275 | 100 | ND | ND | ND | ND | |||||
| Analog 77 | SR-03000002543-1 |
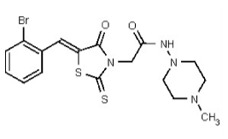 | 53361995 | 125264843 | TSRI | None | ND | 50 | 4X | ND | ND | ND | ||||
| Analog 78 | SR-03000002544-1 |
 | 53361997 | 125264844 | TSRI | None | ND | >50 | ND | ND | ND | |||||
| Analog 79 | SR-03000002545-1 |
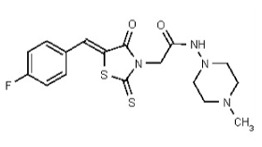 | 53362020 | 125264845 | TSRI | None | ND | >50 | ND | ND | ND | |||||
| Analog 80 | SR-03000002546-1 |
 | 53362005 | 125264846 | TSRI | None | ND | >50 | ND | ND | ND | |||||
| Analog 81 | SR-03000002547-1 |
 | 53362003 | 125264847 | TSRI | None | ND | 50 | 2X | ND | ND | ND | ||||
| Analog 82 | SR-03000002548-1 |
 | 53362016 | 125264848 | TSRI | None | ND | 25 | 4X | ND | ND | ND | ||||
| Analog 83 | SR-03000002549-1 |
 | 53362008 | 125264849 | TSRI | None | ND | 50 | 4X | ND | ND | ND | ||||
| Analog 84 | SR-03000002550-1 |
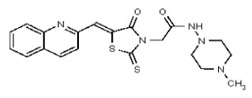 | 53362009 | 125264850 | TSRI | None | ND | >50 | ND | ND | ND | |||||
| Analog 85 | SR-03000002551-1 |
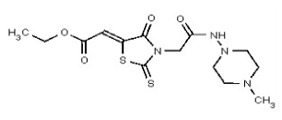 | 53362004 | 125264851 | TSRI | None | ND | 50 | 2X | ND | ND | ND | ||||
| Analog 86 | SR-03000002552-1 |
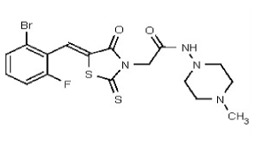 | 53362006 | 125264852 | TSRI | None | ND | 50 | 2X | ND | ND | ND | ||||
| Analog 87 | SR-03000002553-1 |
 | 53362014 | 125264853 | TSRI | None | ND | 25 | 4X | ND | ND | ND | ||||
| Analog 88 | SR-03000002554-1 |
 | 53362019 | 125264854 | TSRI | None | 12.5 | 3.125 | 32X | >50 | 2X | >50 | >50 | |||
| Analog 89 | SR-03000002556-1 |
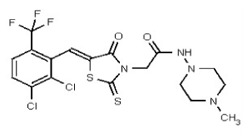 | 53362002 | 125264856 | TSRI | None | 1.563 | 8X | 3.125 | 32X | >50 | 2X | 50 | 4X | >50 | |
| Analog 90 | SR-03000002557-1 |
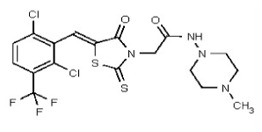 | 53362007 | 125264857 | TSRI | None | 0.781 | 8X | 3.125 | 16X | >50 | 2X | >50 | 2X | >50 | |
| Analog 91 | SR-03000002587-1 |
 | 53384696 | 125311271 | TSRI | None | 25 | 8X | 3.125 | 2X | >50 | 2X | >50 | >50 | ||
| Analog 92 | SR-03000002673-1 |
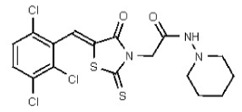 | 53495084 | 126723254 | TSRI | None | ND | 3.125 | 16X | >50 | >50 | >50 | ||||
| Analog 93 | SR-03000002706-1 |
 | 54579793 | 131269028 | TSRI | None | ND | 6.25 | 16X | ND | ND | ND | ||||
| Analog 94 | SR-03000002707-1 |
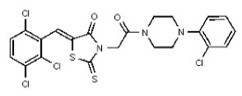 | 54579790 | 131269029 | TSRI | None | ND | >50 | ND | ND | ND | |||||
| Analog 95 | SR-03000002806-1 |
 | 56596523 | 134228483 | TSRI | None | ND | 1.563 | 32X | ND | ND | ND | ||||
| Analog 96 | SR-03000002807-1 |
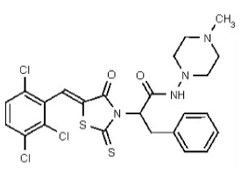 | 56596515 | 134228484 | TSRI | None | ND | 1.563 | 32X | ND | ND | ND | ||||
| Analog 97 | SR-03000002808-1 |
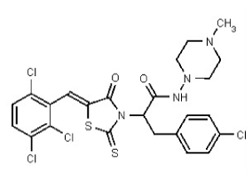 | 56596526 | 134228485 | TSRI | None | ND | 1.563 | 32X | ND | ND | ND | ||||
3.5. Cellular Assays
The probe was tested in a variety of bacterial assays performed by the assay provider to determine the probe’s selectivity, efficacy and mechanism of action.
MIC and Imipenem Antibiotic Synergy Assays [AID 624081 BL21 (VIM2); AID 624097 BL21 (IMP1); AID 624096 YMC07 (VIM2); AID 624082 BAA2146 (NDM1); AID 624080 PA641 (VIM2); AID 624095 KN20 (IMP1)]. Using a checkerboard microdilution method, probe ML302 was tested for its ability to potentiate the efficacy of imipenem on VIM-2-expressing E. coli (BL21/VIM-2) [20–22]. The probe exhibited synergy with imipenem when present at concentrations as low as 3.13 μM, potentiating imipenem’s activity 8-fold. Significantly, probe ML302 exhibited 32-fold imipenem potentiation in clinically relevant VIM-2-transformed Acinetobacter species YMC07/B3323: MIC range = 390–781 nM). Synergy was also observed by probe ML302 in other clinical isolates including K. pneumonia BAA-2146 expressing NDM-1 [2-fold], P. aeruginosa expressing VIM-2 (PA641) [4-fold], and P. aeruginosa expressing IMP-1 (KN20) [2-fold].
3.6. Profiling Assays
The identified VIM-2/IMP-1 inhibitor probe ML302 was tested in biochemical and bacterial MIC assays to determine inhibition of VIM-2, IMP-1, TEM-1, AmpC, as well as assess its ability to potentiate the antibacterial activity of imipenem. Based on the results of these assays, it was agreed by the SRIMSC and assay provider that additional profiling was not required at this time.
4. Discussion
4.1. Comparison to existing art and how the new probe is an improvement
Compared to our prior probe ML121 and prior art mitoxantrone, the new probe ML302 exhibits improved biochemical potency, antibacterial activity, and IMP-1 activity.
Table 4Comparison of ML302 to existing Art
| Name | Structure | SR Number | CID | SID | MLS ID | VIM-2 IC50 | IMP-1 IC50 | VIM-2 Ki | TEM-1 IC50 | AmpC IC50 | Synergy | PubChem Activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe ML302 |
 | SR-03000002555-2 | 53362017 | 134220672 | MLS- 003940491 | 548 nM | 3.02 μM | 183 nM | >60 μM | >60 μM | 3.125 μM (VIM-2) 0.781 μM (IMP-1) (SID 125264855) | None |
| Probe ML121 |
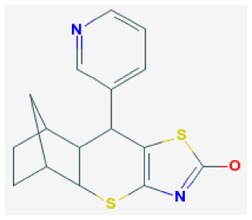 | SR-01000775688-1 | 4870494 | 24790728 (MLSMR) ;85856282 (purch); 103911139 (synth) | MLS-000680027 | 223 nM | >60 μM | 148 nM | >60 μM | ND | Active (12.5 μM) (TFA salt) | 6/309 (1.9%) |
| Prior art Mitoxantrone |
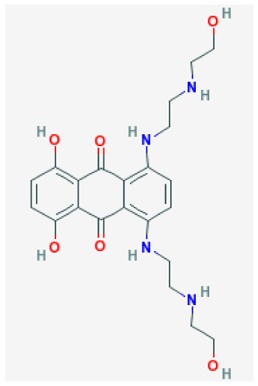 | SR-01000076001-7 | 4212(5458171) | 56424031 (85856281) | MLS 001333711 | powder 0.63 μM | > 56 μM | 1.5 μM (non-competitive): | > 56 μM | ND | Not applicable | 40/139 (28.8%) |
4.2. Mechanism of Action Studies
The assay provider has performed assays to elucidate the mode of inhibition and the probe’s inhibitory constant (Ki) against VIM-2 and IMP-1. The results of these probe development efforts determined that probe ML302 is a non-selective, mixed mode inhibitor probe of the metallo-beta-lactamases VIM-2 and IMP-1 (Figure 6).
VIM-2 and IMP-1 Ki Assays (AID 624083 andAID 624084). The purpose of these assays is to determine the inhibition constant (Ki) and modality of probe candidate molecules. Kinetic assays were conducted by incubating a range of nitrocefin substrate concentrations (100nM - 5 μM) with varying inhibitor concentrations and 0.1 nM enzyme at room temperature in buffer containing 50mM HEPES, 50μM ZnSO4, 0.05% Brij 35, pH 7.1. Absorbance was measured on a Tecan Safire2 monochromatic microplate reader at 495 nm. Initial velocities were obtained from plots of absorbance at 495 nm versus time, using data points from only the linear portion of the hydrolysis curve. Substrate hydrolysis was continuously monitored. Initial velocities were plotted vs. substrate concentration and kinetic parameters were calculated using Graphpad Prism version 5.01 suite of programs. Mode of inhibition was determined using fit comparison capability of Graphpad Prism version 5.01 and additionally evaluated by Lineweaver-Burke plot. Ki values were determined by non-linear regression analysis. The results of these studies demonstrated that probe ML302 (CID 53362017) is a mixed mode uncompetitive and non-competitive inhibitor, with a submicromolar VIM-2 (183 ± 24 nM) and IMP-1 (930 ± 97 nM) Ki values (Figure 6).
5. References
- 1.
- Mazerski J, Martelli S, Borowski E. The geometry of intercalation complex of antitumor mitoxantrone and ametantrone with DNA: molecular dynamics simulations. Acta Biochim Pol. 1998;45(1):1–11. [PubMed: 9701490]
- 2.
- Minond D, Saldanha SA, Subramaniam P, Spaargaren M, Spicer T, Fotsing JR, Weide T, Fokin VV, Sharpless KB, Galleni M, Bebrone C, Lassaux P, Hodder P. Inhibitors of VIM-2 by screening pharmacologically active and click-chemistry compound libraries. Bioorg Med Chem. 2009;17(14):5027–37. [PMC free article: PMC2759276] [PubMed: 19553129]
- 3.
- Siegel RE. Emerging gram-negative antibiotic resistance: daunting challenges, declining sensitivities, and dire consequences. Respir Care. 2008;53(4):471–9. [PubMed: 18364060]
- 4.
- Gupta V. An update on newer beta-lactamases. Indian J Med Res. 2007;126(5):417–27. [PubMed: 18160745]
- 5.
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–51. table of contents. [PMC free article: PMC89009] [PubMed: 11585791]
- 6.
- Sacha P, Wieczorek P, Hauschild T, Zorawski M, Olszanska D, Tryniszewska E. Metallo-beta-lactamases of Pseudomonas aeruginosa--a novel mechanism resistance to beta-lactam antibiotics. Folia Histochem Cytobiol. 2008;46(2):137–42. [PubMed: 18519228]
- 7.
- Koch AL. Bacterial wall as target for attack: past, present, and future research. Clin Microbiol Rev. 2003;16(4):673–87. [PMC free article: PMC207114] [PubMed: 14557293]
- 8.
- Jin W, Arakawa Y, Yasuzawa H, Taki T, Hashiguchi R, Mitsutani K, Shoga A, Yamaguchi Y, Kurosaki H, Shibata N, Ohta M, Goto M. Comparative study of the inhibition of metallo-beta-lactamases (IMP-1 and VIM-2) by thiol compounds that contain a hydrophobic group. Biol Pharm Bull. 2004;27(6):851–6. [PubMed: 15187432]
- 9.
- Abeylath SC, Turos E. Drug delivery approaches to overcome bacterial resistance to beta-lactam antibiotics. Expert Opin Drug Deliv. 2008;5(9):931–49. [PubMed: 18754746]
- 10.
- Wang Z, Fast W, Valentine AM, Benkovic SJ. Metallo-beta-lactamase: structure and mechanism. Curr Opin Chem Biol. 1999;3(5):614–22. [PubMed: 10508665]
- 11.
- Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18(2):306–25. [PMC free article: PMC1082798] [PubMed: 15831827]
- 12.
- Hirakata Y, Izumikawa K, Yamaguchi T, Takemura H, Tanaka H, Yoshida R, Matsuda J, Nakano M, Tomono K, Maesaki S, Kaku M, Yamada Y, Kamihira S, Kohno S. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-beta-lactamase gene blaIMP. Antimicrob Agents Chemother. 1998;42(8):2006–11. [PMC free article: PMC105724] [PubMed: 9687398]
- 13.
- Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43(7):1584–90. [PMC free article: PMC89328] [PubMed: 10390207]
- 14.
- Wang CX, Mi ZH. Imipenem-resistant Pseudomonas aeruginosa producing IMP-1 metallo-beta-lactamases and lacking the outer-membrane protein OprD. J Med Microbiol. 2006;55(Pt 3):353–4. [PubMed: 16476803]
- 15.
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–86. [PMC free article: PMC1265908] [PubMed: 16223952]
- 16.
- Zuck P, O’Donnell GT, Cassaday J, Chase P, Hodder P, Strulovici B, Ferrer M. Miniaturization of absorbance assays using the fluorescent properties of white microplates. Anal Biochem. 2005;342(2):254–9. [PubMed: 15949786]
- 17.
- Li X, He Y, Ruiz CH, Koenig M, Cameron MD, Vojkovsky T. Characterization of dasatinib and its structural analogs as CYP3A4 mechanism-based inactivators and the proposed bioactivation pathways. Drug Metab Dispos. 2009;37(6):1242–50. [PMC free article: PMC3202349] [PubMed: 19282395]
- 18.
- Li X, Kamenecka TM, Cameron MD. Bioactivation of the epidermal growth factor receptor inhibitor gefitinib: implications for pulmonary and hepatic toxicities. Chem Res Toxicol. 2009;22(10):1736–42. [PubMed: 19803472]
- 19.
- Giles RG, LN, Quick JK, Sasse MJ, Urquhart MWJ, Youssef L. Regiospecific Reduction of 5-Benzylidene-2,4-Thiazolidinediones and 4-Oxo-2-thiazolidinethiones using Lithium Borohydride in Pyridine and Tetrahydrofuran. Tetrahedron. 2000;56(26):4531–4537.
- 20.
- Bonapace CR, Bosso JA, Friedrich LV, White RL. Comparison of methods of interpretation of checkerboard synergy testing. Diagn Microbiol Infect Dis. 2002;44(4):363–6. [PubMed: 12543542]
- 21.
- Bajaksouzian S, Visalli MA, Jacobs MR, Appelbaum PC. Activities of levofloxacin, ofloxacin, and ciprofloxacin, alone and in combination with amikacin, against acinetobacters as determined by checkerboard and time-kill studies. Antimicrob Agents Chemother. 1997;41(5):1073–6. [PMC free article: PMC163853] [PubMed: 9145872]
- 22.
- Wayne. E. C. a. L. S. Institute, editor. M26-A: Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline. National Committee on Clinical Laboratory Standards; 1999.
Figures

Figure 1Stability Analysis of ML302 and its HCl salt

Figure 2Synthesis of VIM-2 Inhibitor Probe CID 53362017 (ML302) (Compound 2)

Figure 3Probe analogs