NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
The recently discovered apelin receptor (APJ, AGTRL-1, APLNR) system has emerged as a critical mediator of cardiovascular homeostasis and is involved in the pathogenesis of hypertension, heart failure, atherosclerosis, and other cardiovascular diseases. We disclose the first discovery and characterization of a potent (1.7 – 2.2 μM), kojic acid based small molecule APJ functional antagonist in cell-based assays that is >37 fold selective over the closely related angiotensin II type 1 (AT1) receptor, derived from an high throughput screening (HTS) of the ~330,600 compound MLSMR collection. This antagonist showed no significant binding activity against 29 other GPCRs, except to the κ-opioid receptor (<50%I at 10 μM). The synthetic methodology, development of structure-activity relationship (SAR), and initial in vitro pharmacologic characterization are also presented. This probe molecule provides a useful tool compound for investigators interested in understanding apelin receptor pharmacology and function.
Assigned Assay Grant #: 1R21NS059422-01
Screening Center Name & PI: Sanford-Burnham Center for Chemical Genomics (BCCG) & John C. Reed (PI)
Chemistry Center Name & PI: Sanford-Burnham Center for Chemical Genomics (BCCG) & John C. Reed (PI)
Assay Submitter & Institution: Layton H. Smith, Sanford-Burnham Medical Research Institute
PubChem Summary Bioassay Identifier (AID): 2569
Probe Structure & Characteristics
This Center Probe Report describes the first reported small molecule APJ antagonist, ML221 that is also selective against the AT1 receptor and cell active

Table 1Biological activity summary & AIDs for Probe
| CID/ML# | Target Name | IC50 (nM) [SID, AID] | Anti-target Name(s) | IC50 (μM) [SID, AID] | Fold Selective |
|---|---|---|---|---|---|
| CID 7217941 ML221 | APJ Receptor | 1750 nM SID 103061721 AID 492986 | AT1 Receptor | >79 μM SID 103061721 AID 492984 | >45 |
1. Recommendations for Scientific Use of the Probe
The recently discovered apelin system is emerging as a critical mediator of cardiovascular homeostasis, whose importance in the pathogenesis of hypertension, heart failure, atherosclerosis, and other cardiovascular diseases, is the subject of intense investigation1–7. These investigations are limited by the paucity of research tools and reagents needed to fully understand the role of apelin in physiology and pathology. One particular challenge is the lack of a robust apelin receptor deficient (knock-out) mouse. Although this mouse has been created and reported in the literature, it is difficult to breed homozygous, apelin receptor null mice. Therefore a small molecule antagonist of the apelin receptor (a.k.a APJ, AGTRL-1, APLNR) would advance apelin research significantly. While not a replacement of a transgenic apelin receptor deficient mouse, a small molecule antagonist would be a useful tool for understanding the pharmacology of APJ and ultimately to validate the importance of this system in animal models of cardiovascular disease. To date, there are no small antagonists for the apelin receptor. In this report we disclose the discovery and characterization of a potent selective apelin receptor antagonist. Synthetic methodology, SAR, and activity of our most efficacious and selective compound in a cell-based assay of apelin receptor inhibition are presented. The probe molecule can be used as a tool compound by investigators interested in understanding apelin receptor pharmacology and function.
2. Materials and Methods
2.1. Assays
Table 2 summarizes the details for the assays that drove this probe project and can be found in the “Assay Description” section under the listed AIDs in Table 2 as viewable in PubChem Bioassays.
Table 2
Summary of Assays and AIDs.
All assays in Table 2, with the exception of the β-galactosidase counterscreen (AID 485352), utilize the DiscoveRx PathHunter® β-arrestin assay technology. Unlike imaging or other second messenger assays, the DiscoveRx β-arrestin assay allows for a direct measure of GPCR activation by detection of β-arrestin binding to the APJ, and in the case of the counterscreen, the AT1 receptor. In this system, β-arrestin is fused to an N-terminal deletion mutant of β-galactosidase (termed the enzyme acceptor of EA) and the GPCR of interest is fused to a smaller (42 amino acids), weakly complementing fragment termed ProLink™. In cells that stably express these fusion proteins, ligand stimulation results in the interaction of β-arrestin and the ProLink-tagged GPCR, forcing the complementation of the two β-galactosidase fragments and resulting in the formation of a functional enzyme that converts substrate to detectable luminescent signal.
2.2. Probe Chemical Characterization
Chemical name of probe compound & structure including stereochemistry
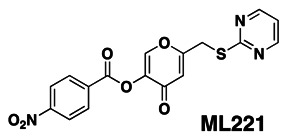
The IUPAC name of the probe is 4-oxo-6-((pyrimidin-2-ylthio)methyl)-4H-pyran-3-yl 4-nitrobenzoate. The actual batch prepared, tested and submitted to the MLSMR is archived as SID 103061721 corresponding to CID 7217941. The probe ML221 has no chiral centers (Figure 1). This probe is not commercially available. A 25 mg sample of ML221 synthesized at SBCCG has been deposited in the MLSMR (Bio-Focus DPI).

Figure 1
Chemical structure of ML221.
Synthetic Route is in Scheme 1 and structure proof by 1HNMR & LC-MS is in Figures 2 and 3A and 3B.

Scheme 1
Synthesis of ML221, conditions. a. neat SOCl2 (17 eq); b. pyrimidine-2-thiol (1 eq), NaOMe (1 eq), MeCN; c. 4-nitrobenzoyl chloride (1.4 eq), Cs2CO3 (1 eq), MeCN.

Figure 2
1H NMR Spectra for ML221.

Figure 3A
LC of LC-MS Data for ML221.

Figure 3B
MS of LC-MS Data for ML221.
Solubility and Stability of probe in PBS at room temperature
The stability and solubility of ML221 was investigated in PBS buffer at room temperature (Figure 4). Initial experiments examining the stability of ML221 at its solubility limit suggested that the probe degrades relatively rapidly in PBS buffer. Thus, we subsequently examined the stability of ML221 with 50% acetonitrile/PBS. In 50% acetonitrile/PBS buffer the compound is soluble, and appears to be stable out to 48 hr (92% remaining).
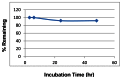
Fig. 4
Stability of ML221 in 1:1 PBS:ACN at ambient temp.
2.3. Probe Preparation
Details of Synthesis and Structural Verification Information of probe SID 103061721 corresponding to CID 7217941
Step 1: A mixture of 5-hydroxy-2-(hydroxymethyl)-4H-pyran-4-one (Kojic acid) (0.55 g, 3.87 mmol) was dissolved in thionyl chloride (5 ml, 68.5 mmol) and was stirred at ambient temperature for 3 hours. Excess reagent was removed in vacuo to provide 0.61 g. (98%) of 2-(chloromethyl)-5-hydroxy-4H-pyran-4-one as an off-white solid. 1H NMR. (500 MHz, DMSO-d6): δ (ppm) 8.13 (s, 1H), 6.57 (s, 1H), 4.66 (s, 2H).
Step 2: A mixture of pyrimidine-2-thiol (161 mg, 1.433 mmol) in 2 ml methanol was treated with sodium methoxide solution (310 mg, 1.433 mmol) and stirred until dissolved. Acetonitrile (10 ml) was added followed by 2-(chloromethyl)-5-hydroxy-4H-pyran-4-one (230 mg, 1.433 mmol) and the mixture was stirred at ambient temperature for 3 hours at which time analysis by LC/MS indicated the reaction to be complete. The solvent was removed in vacuo to provide 406 mg (96%) of a yellow solid containing crude 5-hydroxy-2-((pyrimidin-2-ylthio)methyl)-4H-pyran-4-one and an equimolar amount of sodium chloride which was used without further purification. 1H NMR. (500 MHz, CDCl3): δ (ppm) 8.52 (d, 2H, J= 4.9 Hz), 7.80 (s, 1H), 7.02 (t, 1H, J= 4.8 Hz), 6.63 (s, 1H), 4.23 (s, 2H).
Step 3: A mixture of 5-hydroxy-2-((pyrimidin-2-ylthio)methyl)-4H-pyran-4-one (200 mg, 0.847 mmol), cesium carbonate (276 mg, 0.847 mmol), and 4-nitrobenzoyl chloride (220 mg, 1.185 mmol) in acetonitrile (8 ml) was stirred at ambient temperature overnight. The solvent was removed in vacuo to provide a pale yellow solid, which was partitioned with approximately 20 ml of 1:1 ethyl acetate and water. The desired product remained insoluble in the biphase and was collected by filtration. The solid was dried in vacuo to yield 202 mg (62%) as a tan solid. 1H NMR. (500 MHz, DMSO-d6): δ (ppm) 8.69 (d, 2H, J=4.8 Hz), 8.68 (s, 1H), 8.40 (d, 2H, J=8.8 Hz), 8.29 (d, 2H, J=8.8 Hz), 7.29 (t, 1H, J= 4.9 Hz), 6.65 (s, 1H), 4.45 (s, 2H). 13C NMR. (125 MHz, DMSO-d6): δ (ppm) 171.2, 168.8, 165.9, 161.7, 158.1, 150.8, 149.9, 140.3, 133.0, 131.4, 124.2, 118.0, 114.6, 31.2.
3. Results
3.1. Dose Response Curves for Probe
3.2. Cellular Activity
All of the primary and selectivity assays are cell based.
Table 4Cellular Activity of Probe ML221
| Probe | CID | SID | MLS-# | S/P* | Chemical Structure | Potency (μM)a | ||
|---|---|---|---|---|---|---|---|---|
| APJ | AT1 | SI | ||||||
| ML221 | CID 7217941 | SID 99213078 | 0437359 | P |
 | 2.16 ± 0.96 | >79 | >37 |
| SID 103061721 | S | 1.75 ± 0.19 | >79 | >45 | ||||
- *
S = Synthesized P = purchased
- a
Ave. ± S.E.M. (n = 4), if replicates is different than n=4 it is noted in parentheses (AIDs in Table 1). SI = Selectivity Index: (IC50 AT1)/(IC50 APJ)
3.3. Profiling Assays
The probe was evaluated in a detailed in vitro pharmacology panel as shown in Table 5.
Table 5
Summary of in vitro ADME Properties of APJ Antagonist probe ML221.
ML221 is poorly soluble in aqueous media at pH 5.0/6.2/7.4, although the solubility appears pH-dependent as it is almost three-fold higher in pH 7.4 than in either pH6.2 or pH5.0. We note that the aqueous solubilities obtained at physiological pH are 5 – 14 fold higher than the obtained potency of the probe.
The PAMPA (Parallel Artificial Membrane Permeability Assay) assay is used as an in vitro model of passive, transcellular permeability. An artificial membrane immobilized on a filter is placed between a donor and acceptor compartment. At the start of the test, drug is introduced in the donor compartment. Following the permeation period, the concentration of drug in the donor and acceptor compartments is measured using UV spectroscopy. Consistent with its solubility data, ML221 exhibits moderate permeability that increased with pH.
Plasma protein binding is a measure of a drug’s efficiency to bind to the proteins within blood plasma. The less bound a drug is, the more efficiently it can traverse cell membranes or diffuse. Highly plasma protein bound drugs are confined to the vascular space, thereby having a relatively low volume of distribution. In contrast, drugs that remain largely unbound in plasma are generally available for distribution to other organs and tissues. ML221 was undetectable in the plasma protein binding assay indicating that the compound is likely rapidly metabolized in plasma.
Plasma stability is a measure of the stability of small molecules and peptides in plasma and is an important parameter, which strongly can influence the in vivo efficacy of a test compound. Drug candidates are exposed in plasma to enzymatic processes (proteinases, esterases), and they can undergo intramolecular rearrangement or bind irreversibly (covalently) to proteins. ML221 shows very poor stability in human plasma (<1% remaining) after 3 hr. This data explains the lack of compound detected in the plasma protein binding assay, as that assay involves an 18h incubation.
The microsomal stability assay is commonly used to rank compounds according to their metabolic stability. This assay addresses the pharmacologic question of how long the parent compound will remain circulating in plasma within the body. ML221 shows poor stability (4.2 % & 4.9% remaining at 60 min) in both human and mouse liver homogenates. Neither the plasma nor the microsomal stability assay results are surprising given the ester linkage in this probe. Attempts to replace the ester with a more stable functional group resulted in inactive compounds, although replacements were not widely investigated. Ultimately this limits the utility of this probe to in vitro studies or apelin receptor or in vivo studies using acute intravenous doses to avoid metabolism.
ML221 shows no toxicity (>50 μM) toward human hepatocytes.
Profiling against other GPCRs. The probe, ML221 (CID7217941), was submitted to the Psychoactive Drug Screening Program (PDSP) at the University of North Carolina (PDSP, Bryan Roth, PI) and the data against a GPCR binding assay panel is shown in Figure 6. Overall, the compound shows a relatively clean binding profile with the only significant activity at the kappa opioid receptor.

Figure 6
GPCR profiling panel for ML221.
4. Discussion
Currently there are no small molecule tools to investigate the biological functions of apelin and its receptor. Apelin is the endogenous peptide ligand8 for the G-protein coupled receptor (GPCR) APJ (angiotensin II receptor-like 1, AGTRL-1 and APLNR). Until the discovery of apelin, APJ was an orphan GPCR. APJ is coupled to Gαi, and has been shown in cell culture to inhibit adenylate cyclase. The APJ gene encodes a receptor that most closely resembles the angiotensin receptor AT1. However, the APJ receptor does not bind angiotensin II9. Underscoring the emerging importance of the apelin/APJ system, recent studies have shown that apelin reduces the extent of atherosclerotic lesions in ApoE−/− mice (7), and opposes the development of abdominal aortic aneurysms10. Additionally, work in Dr. Smith’s lab has revealed that APJ forms a heterodimer with the Ang II receptor AT111, and that this complex facilitates antagonism of Ang II signaling by apelin. Despite these exciting results, there remains a multitude of unanswered questions regarding the role of apelin and APJ in the physiology and pathology, so chemical biological tools would be an advance.
4.1. Comparison to Existing Art and How the New Probe is an Improvement
There are no reports of APJ antagonists in the literature, including in the patent literature. Thus, ML221 represents the first tool compound to probe APJ receptor biology. A recent SciFinder search (March 14, 2011) around the scaffold (holding the benzoate ester and thiopyrimidine constant) shows that it is not widely cited and thus is unlikely to be a promiscuous compound.
5. References
- 1.
- De Falco M, De Luca L, Onori N, Cavallotti I, Artigiano F, Esposito V, De Luca B, Laforgia V, Groeger AM, De Luca A. Apelin expression in normal human tissues. In Vivo. 2002;16:333–336. [PubMed: 12494873]
- 2.
- Kleinz MJ, Davenport AP. Immunocytochemical localization of the endogenous vasoactive peptide apelin to human vascular and endocardial endothelial cells. Regul Pept. 2004;118:119–125. [PubMed: 15003827]
- 3.
- Kleinz MJ, Skepper JN, Davenport AP. Immunocytochemical localization of the apelin receptor, APJ, to human cardiomyocytes, vascular smooth muscle and endothelial cells. Regul Pept. 2005;126:233–240. [PubMed: 15664671]
- 4.
- Quazi R, Palaniswamy C, Frishman WH. The emerging role of apelin in cardiovascular disease and health. Cardiol Rev. 2009;17:283–286. [PubMed: 19829178]
- 5.
- Sorli SC, van den Berghe L, Masri B, Knibiehler B, Audigier Y. Therapeutic potential of interfering with apelin signaling. Drug Discov Today. 2006;11:1100–1106. [PubMed: 17129829]
- 6.
- Ashley EA, Powers J, Chen M, Kundu R, Finsterbach T, Caffarelli A, Deng A, Eichhorn J, Mahajan R, Agrawal R, et al. The endogenous peptide apelin potently improves cardiac contractility and reduces cardiac loading in vivo. Cardiovasc Res. 2005;65:73–82. [PMC free article: PMC2517138] [PubMed: 15621035]
- 7.
- Chun HJ, Ali ZA, Kojima Y, Kundu RK, Sheikh AY, Agrawal R, Zheng L, Leeper NJ, Pearl NE, Patterson AJ, et al. Apelin signaling antagonizes Ang II effects in mouse models of atherosclerosis. J Clin Invest. 2008;118:3343–3354. [PMC free article: PMC2525695] [PubMed: 18769630]
- 8.
- Lee DK, Cheng R, Nguyen T, Fan T, Kariyawasam AP, Liu Y, Osmond DH, George SR, O’Dowd BF. Characterization of apelin, the ligand for the APJ receptor. J Neurochem. 2000;74:34–41. [PubMed: 10617103]
- 9.
- O’Dowd BF, Heiber MM, Chan AA, Heng HHH, Tsui LLC, Kennedy JJL, Shi XX, Petronis AA, George SSR, Nguyen TT. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene. 1993;136:355. [PubMed: 8294032]
- 10.
- Leeper NJ, Tedesco MM, Kojima Y, Schultz GM, Kundu RK, Ashley EA, Tsao PS, Dalman RL, Quertermous T. Apelin prevents aortic aneurysm formation by inhibiting macrophage inflammation. Am J Physiol Heart Circ Physiol. 2009;296:H1329–1335. [PMC free article: PMC2685356] [PubMed: 19304942]
- 11.
- Lee DK, Lanca AJ, Cheng R, Nguyen T, Ji XD, Gobeil F Jr, Chemtob S, George SR, O’Dowd BF. Agonist-independent nuclear localization of the Apelin, angiotensin AT1, and bradykinin B2 receptors. J Biol Chem. 2004;279:7901–7908. [PubMed: 14645236]
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Functional Agonists of the Apelin (APJ) Receptor.[Probe Reports from the NIH Mol...]Review Functional Agonists of the Apelin (APJ) Receptor.Khan P, Maloney PR, Hedrick M, Gosalia P, Milewski M, Li L, Roth GP, Sergienko E, Suyama E, Sugarman E, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Discovery of 4-oxo-6-((pyrimidin-2-ylthio)methyl)-4H-pyran-3-yl 4-nitrobenzoate (ML221) as a functional antagonist of the apelin (APJ) receptor.[Bioorg Med Chem Lett. 2012]Discovery of 4-oxo-6-((pyrimidin-2-ylthio)methyl)-4H-pyran-3-yl 4-nitrobenzoate (ML221) as a functional antagonist of the apelin (APJ) receptor.Maloney PR, Khan P, Hedrick M, Gosalia P, Milewski M, Li L, Roth GP, Sergienko E, Suyama E, Sugarman E, et al. Bioorg Med Chem Lett. 2012 Nov 1; 22(21):6656-60. Epub 2012 Sep 7.
- Discovery of a novel small molecule agonist scaffold for the APJ receptor.[Bioorg Med Chem. 2016]Discovery of a novel small molecule agonist scaffold for the APJ receptor.Narayanan S, Maitra R, Deschamps JR, Bortoff K, Thomas JB, Zhang Y, Warner K, Vasukuttan V, Decker A, Runyon SP. Bioorg Med Chem. 2016 Aug 15; 24(16):3758-70. Epub 2016 Jun 11.
- Review Apelin and its receptor APJ in cardiovascular diseases.[Clin Chim Acta. 2014]Review Apelin and its receptor APJ in cardiovascular diseases.Yu XH, Tang ZB, Liu LJ, Qian H, Tang SL, Zhang DW, Tian GP, Tang CK. Clin Chim Acta. 2014 Jan 20; 428:1-8. Epub 2013 Sep 18.
- Review Apelin: a novel neurohumoral modulator of the cardiovascular system. Pathophysiologic importance and potential use as a therapeutic target.[Rev Port Cardiol. 2005]Review Apelin: a novel neurohumoral modulator of the cardiovascular system. Pathophysiologic importance and potential use as a therapeutic target.Falcão-Pires I, Leite-Moreira AF. Rev Port Cardiol. 2005 Oct; 24(10):1263-76.
- Functional antagonists of the Apelin (APJ) receptor - Probe Reports from the NIH...Functional antagonists of the Apelin (APJ) receptor - Probe Reports from the NIH Molecular Libraries Program
- RecName: Full=Major mite allergen Der p 23; AltName: Full=Major house dust mite ...RecName: Full=Major mite allergen Der p 23; AltName: Full=Major house dust mite allergen Der p 23; Short=Major HDM allergen Der p 23; AltName: Full=Peritrophin-like protein Der p 23; AltName: Allergen=Der p 23; Flags: Precursorgi|1559988684|sp|L7N6F8.1|DEP23_DERProtein
Your browsing activity is empty.
Activity recording is turned off.
See more...

