By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Gilbert SF. Developmental Biology. 6th edition. Sunderland (MA): Sinauer Associates; 2000.

Developmental Biology. 6th edition.
Show detailsThe anterior-posterior polarity of the embryo, larva, and adult has its origin in the anterior-posterior polarity of the egg (Figure 9.8). The maternal effect genes expressed in the mother's ovaries produce messenger RNAs that are placed in different regions of the egg. These messages encode transcriptional and translational regulatory proteins that diffuse through the syncytial blastoderm and activate or repress the expression of certain zygotic genes. Two of these proteins, Bicoid and Hunchback, regulate the production of anterior structures, while another pair of maternally specified proteins, Nanos and Caudal, regulates the formation of the posterior parts of the embryo. Next, the zygotic genes regulated by these maternal factors are expressed in certain broad (about three segments wide), partially overlapping domains. These genes are called gap genes (because mutations in them cause gaps in the segmentation pattern), and they are among the first genes transcribed in the embryo. Differing concentrations of the gap gene proteins cause the transcription of pair-rule genes, which divide the embryo into periodic units. The transcription of the different pair-rule genes results in a striped pattern of seven vertical bands perpendicular to the anterior-posterior axis. The pair-rule gene proteins activate the transcription of the segment polarity genes, whose mRNA and protein products divide the embryo into 14 segment-wide units, establishing the periodicity of the embryo. At the same time, the protein products of the gap, pair-rule, and segment polarity genes interact to regulate another class of genes, the homeotic selector genes, whose transcription determines the developmental fate of each segment.

Figure 9.8
Generalized model of Drosophila anterior-posterior pattern formation. (A) The pattern is established by maternal effect genes that form gradients and regions of morphogenetic proteins. These morphogenetic determinants create a gradient of Hunchback protein (more...)
The Maternal Effect Genes
Embryological evidence of polarity regulation by oocyte cytoplasm
Classic embryological experiments demonstrated that there are at least two “organizing centers” in the insect egg, one in the anterior of the egg and one in the posterior. For instance, Klaus Sander (1975) found that if he ligated the egg early in development, separating the anterior from the posterior region, one half developed into an anterior embryo and one half developed into a posterior embryo, but neither half contained the middle segments of the embryo. The later in development the ligature was made, the fewer middle segments were missing. Thus, it appeared that there were indeed gradients emanating from the two poles during cleavage, and that these gradients interacted to produce the positional information determining the identity of each segment. Moreover, when the RNA of the anterior of insect eggs was destroyed (by either ultraviolet light or RNase), the resulting embryos lacked a head and thorax. Instead, these embryos developed two abdomens and telsons (tails) with mirror-image symmetry: telson-abdomen-abdomen-telson (Figure 9.9; Kalthoff and Sander 1968; Kandler-Singer and Kalthoff 1976). Thus, Sander's laboratory postulated the existence of a gradient at both ends of the egg, and hypothesized that the egg sequesters an RNA that generates a gradient of anterior-forming material.

Figure 9.9
Normal and irradiated embryos of the midge Smittia. The normal embryo (top) shows a head on the left and abdominal segments on the right. The UV-irradiated embryo has no head region, but has abdominal segments at both ends. (From Kalthoff 1969; photographs (more...)
WEBSITE
9.4 Evidence for gradients in insect development. The original evidence for gradients in insect development came from studies providing evidence for two “organization centers” in the egg, one located anteriorly and one located posteriorly. http://www.devbio.com/chap09/link0904.shtml
The molecular model: protein gradients in the early embryo
In the late 1980s, the gradient hypothesis was united with a genetic approach to the study of Drosophila embryogenesis. If there were gradients, what were the morphogens whose concentrations changed over space? What were the genes that shaped these gradients? And did these morphogens act by activating or inhibiting certain genes in the areas where they were concentrated? Christiane Nüsslein-Volhard led a research program that addressed these questions. The researchers found found that one set of genes encoded gradient morphogens for the anterior part of the embryo, another set of genes encoded morphogens responsible for organizing the posterior region of the embryo, and a third set of genes encoded proteins that produced the terminal regions at both ends of the embryo (Figure 9.10; Table 9.1). This work resulted in a Nobel Prize for Nüsslein-Volhard and her colleague, Eric Wieschaus, in 1995.

Figure 9.10
Three independent genetic pathways interact to form the anterior-posterior axis of the Drosophila embryo. In each case, the initial asymmetry is established during oogenesis, and the pattern is organized by maternal proteins soon after fertilization. (more...)
Table 9.1
Maternal effect genes that effect the anterior-posterior polarity of the Drosophila embryo.
WEBSITE
9.5 Christiane Nüsslein-Volhard and the molecular approach to development. The research that revolutionized developmental biology had to wait for someone to synthesize molecular biology, embryology, and Drosophila genetics. http://www.devbio.com/chap09/link0905.shtml
The anterior-posterior axis of the Drosophila embryo appears to be patterned before the nuclei even begin to function. The nurse cells of the ovary deposit mRNAs in the developing oocyte, and these mRNAs are apportioned to different regions of the cell. In particular, four maternal messenger RNAs are critical to the formation of the anterior-posterior axis:
- bicoid and hunchback mRNAs, whose protein products are critical for head and thorax formation
- nanos and caudal mRNAs, whose protein products are critical for the formation of the abdominal segments
The bicoid mRNAs are located in the anterior portion of the unfertilized egg, and are tethered to the anterior microtubules. The nanos messages are bound to the cytoskeleton in the posterior region of the unfertilized egg. The hunchback and caudal mRNAs are distributed throughout the oocyte. Upon fertilization, these mRNAs can be translated into proteins. At the anterior pole, the bicoid RNA is translated into Bicoid protein, which forms a gradient highest at the anterior. At the posterior pole, the nanos message is translated into Nanos protein, which forms a gradient highest at the posterior. Bicoid protein inhibits the translation of the caudal RNA, allowing Caudal protein to be synthesized only in the posterior of the cell. Conversely, Nanos protein, in conjunction with Pumilio protein, binds to hunchback RNA, preventing its translation in the posterior portion of the embryo. Bicoid also elevates the level of Hunchback protein in the anterior of the embryo by binding to the enhancers of the hunchback gene and stimulating its transcription. The result of these interactions is the creation of four protein gradients in the early embryo (Figure 9.11):
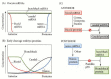
Figure 9.11
A model of anterior-posterior pattern generation by the Drosophila maternal effect genes. (A) The bicoid, nanos, hunchback, and caudal messenger RNAs are placed in the oocyte by the ovarian nurse cells. The bicoid message is sequestered anteriorly. The (more...)
- An anterior-to-posterior gradient of Bicoid protein
- An anterior-to-posterior gradient of Hunchback protein
- A posterior-to-anterior gradient of Nanos protein
- A posterior-to-anterior gradient of Caudal protein
The Bicoid, Hunchback, and Caudal proteins are transcription factors whose relative concentrations can activate or repress particular zygotic genes. The stage is now set for the activation of zygotic genes in those nuclei that were busily dividing while this gradient was being established.
Evidence that the bicoid gradient constitutes the anterior organizing center
In Drosophila, the phenotype of the bicoid mutant provides valuable information about the function of gradients. Instead of having anterior structures (acron, head, and thorax) followed by abdominal structures and a telson, the structure of the bicoid mutant is telson-abdomen-abdomen-telson (Figure 9.12). It would appear that these embryos lack whatever substances are needed for the formation of anterior structures. Moreover, one could hypothesize that the substance that these mutants lack is the one postulated by Sander and Kalthoff to turn on genes for the anterior structures and turn off genes for the telson structures (compare Figures 9.9 and 9.12).
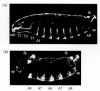
Figure 9.12
Phenotype of a strongly affected embryo from a female deficient in the bicoid gene. (A) Wild-type cuticle pattern. (B) Bicoid mutant. The head and thorax have been replaced by a second set of posterior telson structures. Abbreviations: fk, filzkörper; (more...)
Further studies have strengthened the view that the product of the wild-type bicoid gene is the morphogen that controls anterior development. First, bicoid is a maternal effect gene. Messenger RNA from the mother's bicoid genes is placed in the embryo by the mother's ovarian cells (Figure 9.13A; Frigerio et al. 1986; Berleth et al. 1988). The bicoid RNA is strictly localized in the anterior portion of the oocyte (Figure 9.13B), where the anterior cytoskeleton anchors it through the message's 3´ untranslated region (Ferrandon et al. 1997; Macdonald and Kerr 1998). This mRNA is dormant until fertilization, at which time it receives a longer polyadenylate tail and can be translated. Driever and Nüsslein-Volhard (1988b) have shown that when Bicoid protein is translated from this RNA during early cleavage, it forms a gradient, with the highest concentration in the anterior of the egg and the lowest in the posterior third of the egg. Moreover, this protein soon becomes concentrated in the embryonic nuclei in the anterior portion of the embryo (Figure 9.13C-D; see also Figure 5.35).
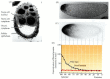
Figure 9.13
Gradient of Bicoid protein in the early Drosophila embryo. (A) bicoid mRNA passing into the oocyte from the nurse cells during oogenesis. (B) Localization of bicoid mRNA to the anterior tip of the embryo. (C) Gradient of Bicoid protein shortly after fertilization. (more...)
WEBSITE
9.6 Mechanism of bicoid mRNA localization. One of the most critical steps in Drosophila pattern formation is the binding of the bicoid mRNA to the anterior microtubules. Several genes are involved in this process, wherein the bicoid message forms a complex with several proteins. http://www.devbio.com/chap09/link0906.shtml
Further evidence that Bicoid protein is the anterior morphogen came from experiments that altered the steepness of the gradient. Two genes, exuperantia and swallow, are responsible for keeping the bicoid message at the anterior pole of the egg. In their absence, the bicoid message diffuses farther into the posterior of the egg, and the gradient of Bicoid protein is less steep (Driever and Nüsslein-Volhard 1988a). The phenotype produced by these two mutants is similar to that of bicoid-deficient embryos, but less severe. These embryos lack their most anterior structures and have an extended mouth and thoracic region. Thus, by altering the gradient of Bicoid protein, one correspondingly alters the fate of the embryonic regions.
Confirmation that the Bicoid protein is crucial for initiating head and thorax formation came from experiments in which purified bicoid RNA was injected into early-cleavage embryos (Figure 9.14; Driever et al. 1990). When injected into the anterior of bicoid-deficient embryos (whose mothers lacked bicoid genes), the bicoid RNA rescued the embryos and caused them to have normal anterior-posterior polarity. Moreover, any location in an embryo where the bicoid message was injected became the head. If bicoid RNA was injected into the center of an embryo, that middle region became the head, and the regions on either side of it became thorax structures. If a large amount of bicoid RNA was injected into the posterior end of a wild-type embryo (with its own endogenous bicoid message in its anterior pole), two heads emerged, one at either end.

Figure 9.14
Schematic representation of the experiments demonstrating that the bicoid gene encodes the morphogen responsible for head structures in Drosophila. The phenotypes of bicoid-deficient and wild-type embryos are shown at the sides. When bicoid-deficient (more...)
The next question then emerged: How might a gradient in Bicoid protein control the determination of the anterior-posterior axis? Recent evidence suggests that Bicoid acts in two ways to specify the anterior of the Drosophila embryo. First, it acts as a repressor of posterior formation. It does this by binding to and suppressing the translation of caudal RNA, which is found throughout the egg and early embryo. The Caudal protein is critical in specifying the posterior domains of the embryo, and it activates the genes responsible for the invagination of the hindgut (Wu and Lengyel 1998). The Bicoid protein binds to a specific region of the caudal message's 3´ untranslated region, thereby preventing the translation of this message in the anterior section of the embryo (Figure 9.15; Dubnau and Struhl 1996; Rivera-Pomar et al. 1996). This suppression is necessary, for if Caudal protein is made in the anterior, the head and thorax are not properly formed.

Figure 9.15
Gradient of Caudal protein in the syncitial blastoderm of a wild-type Drosophila embryo. The protein (stained darkly) enters the nuclei and helps specify posterior fates. Compare with the complementary gradient of Bicoid protein in Figure 9.13. (From (more...)
Second, the Bicoid protein functions as a transcription factor. Bicoid protein enters the nuclei of early-cleavage embryos, where it activates the hunchback gene. The transcription of hunchback is seen only in the anterior half of the embryo—the region where Bicoid protein is found. Mutants deficient in maternal and zygotic Hunchback protein lack mouthparts and thorax structures. In the late 1980s, two laboratories independently demonstrated that Bicoid protein binds to and activates the hunchback gene (Driever and Nüsslein-Volhard 1989; Struhl et al. 1989). The Hunchback protein derived from the synthesis of new hunchback mRNA joins the Hunchback protein synthesized by the translation of maternal messages in the anterior of the embryo. The Hunchback protein, also a transcription factor, represses abdominal-specific genes, thereby allowing the region of hunchback expression to form the head and thorax.
The Hunchback protein also works with Bicoid in generating the anterior pattern of the embryo. Based on two pieces of evidence, Driever and co-workers (1989) predicted that at least one other anterior gene besides hunchback must be activated by Bicoid. First, deletions of hunchback produce only some of the defects seen in the bicoid mutant phenotype. Second, as we saw in the swallow and exuperantia experiments, only moderate levels of Bicoid protein are needed to activate thorax formation (i.e., hunchback gene expression), but head formation requires higher concentrations. Driever and co-workers (1989) predicted that the promoters of such a head-specific gap gene would have low-affinity binding sites for Bicoid protein. This gene would be activated only at extremely high concentrations of Bicoid protein—that is, near the anterior tip of the embryo. Since then, three gap genes of the head have been discovered that are dependent on very high concentrations of Bicoid protein for their expression (Cohen and Jürgens 1990; Finkelstein and Perrimon 1990; Grossniklaus et al. 1994). The buttonhead, empty spiracles, and orthodenticle genes are needed to specify the progressively anterior regions of the head. In addition to needing high Bicoid levels for activation, these genes also require the presence of Hunchback protein to be transcribed (Simpson-Brose et al. 1994; Reinitz et al. 1995). The Bicoid and Hunchback proteins act synergistically at the enhancers of these “head genes” to promote their transcription.
The posterior organizing center: localizing and activating nanos
The posterior organizing center is defined by the activities of the nanos gene (Lehmann and Nüsslein-Volhard 1991;Wang and Lehmann 1991; Wharton and Struhl 1991). The nanos RNA is produced by the ovarian nurse cells and is transported into the posterior region of the egg (farthest away from the nurse cells). The nanos message is bound to the cytoskeleton in the posterior region of the egg through its 3´ UTR and its association with the products of several other genes (oskar, valois, vasa, staufen, and tudor).* If nanos or any other of these maternal effect genes are absent in the mother, no embryonic abdomen forms (Lehmann and Nüsslein-Volhard 1986; Schüpbach and Wieschaus 1986).
The nanos message is dormant in the unfertilized egg, as it is repressed by the binding of the Smaug protein to its 3´ UTR (Smibert et al. 1996). At fertilization, this repression is removed, and Nanos protein can be synthesized. The Nanos protein forms a gradient that is highest at the posterior end. Nanos functions by inactivating hunchback mRNA translation (Figure 9.16, see also Figure 9.11; Tautz 1988). In the anterior of the cleavage-stage embryo, the hunchback message is bound in its 3´ UTR by the Pumilio protein, and the message can be translated into Hunchback protein. In the posterior of the early embryo, however, the bound Pumilio can be joined by the Nanos protein. Nanos binds to Pumilio and deadenylates the hunchback mRNA, preventing its translation (Barker et al. 1992; Wreden et al. 1997). The hunchback RNA is initially present throughout the embryo, although more can be made from zygotic nuclei if they are activated by Bicoid protein. Thus, the combination of Bicoid and Nanos proteins causes a gradient of Hunchback protein across the egg. The Bicoid protein activates hunchback gene transcription in the anterior part of the embryo, while the Nanos protein inhibits the translation of hunchback RNA in the posterior part of the embryo.
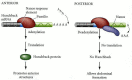
Figure 9.16
Control of hunchback mRNA translation by Nanos. In the anterior of the embryo, Pumilio protein binds to the Nanos Response Element (NRE) in the 3´ UTR of the hunchback message, and the message is polyadenylated normally. This polyadenylated message (more...)
The terminal gene group
In addition to the anterior and posterior morphogens, there is third set of maternal genes whose proteins generate the extremes of the anterior-posterior axis. Mutations in these terminal genes result in the loss of the unsegmented extremities of the organism: the acron and the most anterior head segments and the telson (tail) and the most posterior abdominal segments (Degelmann et al. 1986; Klingler et al. 1988). A critical gene here appears to be torso, a gene encoding a receptor tyrosine kinase. The embryos of mothers with mutations of the torso gene have neither acron nor telson, suggesting that the two termini of the embryo are formed through the same pathway. The torso RNA is synthesized by the ovarian cells, deposited in the oocyte, and translated after fertilization. The transmembrane Torso protein is not spatially restricted to the ends of the egg, but is evenly distributed throughout the plasma membrane (Casanova and Struhl 1989). Indeed, a dominant mutation of torso, which imparts constitutive activity to the receptor, converts the entire anterior half of the embryo into an acron and the entire posterior half into a telson. Thus, Torso must normally be activated only at the ends of the egg.
Stevens and her colleagues (1990) have shown that this is the case. Torso protein is activated by the follicle cells only at the two poles of the oocyte. Two pieces of evidence suggest that the activator of the Torso protein is probably the Torso-like protein: first, loss-of-function mutations in the torso-like gene create a phenotype almost identical to that produced by torso mutants, and second, ectopic expression of Torso-like causes the activation of the Torso protein in the new location. The torso-like gene is usually expressed only in the anterior and posterior follicle cells, and the secreted Torso-like protein can cross the perivitelline space to activate the Torso protein in the egg membrane (Martin et al. 1994; Furriols et al. 1998). In this manner, the Torso-like protein activates the Torso protein in the anterior and posterior regions of the oocyte membrane. The end products of the RTK-kinase cascade activated by the Torso protein diffuse into the cytoplasm at both ends of the embryo (Figure 9.17; Gabay et al. 1997; see Chapter 6). These kinases are thought to inactivate a transcriptional inhibitor of the tailless and huckebein gap genes (Paroush et al. 1997). These two genes then specify the termini of the embryo. The distinction between the anterior and posterior termini depends on the presence of Bicoid. If the terminal genes act alone, the terminal regions differentiate into telsons. However, if Bicoid is also present, the region forms an acron (Pignoni et al. 1992).
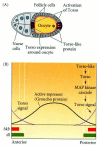
Figure 9.17
Formation of the unsegmented poles by torso signaling. (A) Torso-like protein is expressed by the follicle cells at the poles of the oocyte. Torso protein is expressed around the entire oocyte. Torso-like activates torso protein at the poles (see Casanova (more...)
The anterior-posterior axis of the embryo is therefore specified by three sets of genes: those that define the anterior organizing center, those that define the posterior organizing center, and those that define the terminal boundary region. The anterior organizing center is located at the anterior end of the embryo and acts through a gradient of Bicoid protein that functions as a transcription factor to activate anterior-specific gap genes and as a translational repressor to suppresses posterior-specific gap genes. The posterior organizing center is located at the posterior pole and acts translationally through the Nanos protein to inhibit anterior formation and transcriptionally through the Caudal protein to activate those genes that form the abdomen. The boundaries of the acron and telson are defined by the product of the torso gene, which is activated at the tips of the embryo. The activation of those genes responsible for constructing the posterior is performed by Caudal, a protein whose synthesis (as we have seen above) is inhibited in the anterior portion of the embryo. The next step in development will be to use these gradients of transcription factors to activate specific genes along the anterior-posterior axis.
The Segmentation Genes
The process of cell fate commitment in Drosophila appears to have two steps: specification and determination (Slack 1983). Early in development, the fate of a cell depends on environmental cues, such as those provided by the protein gradients mentioned above. This specification of cell fate is flexible and can still be altered in response to signals from other cells. Eventually, the cells undergo a transition from this loose type of commitment to an irreversible determination. At this point, the fate of a cell becomes cell-intrinsic.† The transition from specification to determination in Drosophila is mediated by the segmentation genes. These genes divide the early embryo into a repeating series of segmental primordia along the anterior-posterior axis. Mutations in segmentation genes cause the embryo to lack certain segments or parts of segments. Often these mutations affect parasegments, regions of the embryo that are separated by mesodermal thickenings and ectodermal grooves. The segmentation genes divide the embryo into 14 parasegments (Martinez-Arias and Lawrence 1985). The parasegments of the embryo do not become the segments of the larva or adult; rather, they include the posterior part of an anterior segment and the anterior portion of the segment behind it (Figure 9.18). While the segments are the major anatomical divisions of the larval and adult body plan, they are built according to rules that use the parasegment as the basic unit of construction.
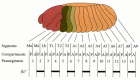
Figure 9.18
Segments and parasegments. A and P represent the anterior and posterior compartments of the segments. The parasegments are shifted one compartment forward. Ma, Mx, and Lb represent three of the head segments (mandibular, maxillary, and labial), the T (more...)
There are three classes of segmentation genes, which are expressed sequentially (see Figure 9.8). The transition from an embryo characterized by gradients of morphogens to an embryo with distinct units is accomplished by the products of the gap genes. The gap genes are activated or repressed by the maternal effect genes, and they divide the embryo into broad regions, each containing several parasegment primordia. The Krüppel gene, for example, is expressed primarily in parasegments 4–6, in the center of the Drosophila embryo (Figures 9.19A; 9.8B); the absence of the Krüppel protein causes the embryo to lack these regions. The protein products of the gap genes interact with neighboring gap gene proteins to activate the transcription of the pair-rule genes. The products of these genes subdivide the broad gap gene regions into parasegments. Mutations of pair-rule genes, such as fushi tarazu (Figures 9.8C, 9.19B, 9.20), usually delete portions of alternate segments. Finally, the segment polarity genes are responsible for maintaining certain repeated structures within each segment. Mutations in these genes cause a portion of each segment to be deleted and replaced by a mirror-image structure of another portion of the segment. For instance, in engrailed mutants, portions of the posterior part of each segment are replaced by duplications of the anterior region of the subsequent segment (Figures 9.19C, 9.8D). Thus, the segmentation genes are transcription factors that use the gradients of the early-cleavage embryo to transform the embryo into a periodic, parasegmental structure.
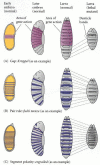
Figure 9.19
Three types of segmentation gene mutations. The left panel shows the early-cleavage embryo, with the region where the particular gene is normally transcribed in wild-type embryos shown in color. These areas are deleted as the mutants develop.
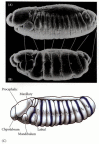
Figure 9.20
Defects seen in the fushi tarazu mutant. (A) Scanning electron micrograph of a wild-type embryo, seen in lateral view. (B) Same stage of a fushi tarazu mutant embryo. The white lines connect the homologous portions of the segmented germ band. (C) Diagram (more...)
After the parasegmental boundaries are set, the pair-rule and gap genes interact to regulate the homeotic selector genes, which determine the identity of each segment. By the end of the cellular blastoderm stage, each segment primordium has been given an individual identity by its unique constellation of gap, pair-rule, and homeotic gene products (Levine and Harding 1989).
The gap genes
The gap genes were originally discovered through a series of mutant embryos that lacked groups of consecutive segments (Figure 9.21; Nüsslein-Volhard and Wieschaus 1980). Deletions caused by mutations of the hunchback, Krüppel, and knirps genes span the entire segmented region of the Drosophila embryo. The giant gap gene overlaps with these three, and mutations of the tailless and huckebein genes delete portions of the unsegmented termini of the embryo.
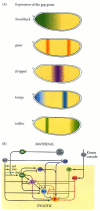
Figure 9.21
Conversion of maternal protein gradients into zygotic gap gene expression. (A) Gap gene expression patterns. (B) The gradients of maternal transcription factors Bicoid, Caudal, and Hunchback regulate the transcription of the gap genes. Hunchback and Caudal (more...)
The expression of the gap genes is dynamic. There is usually a low level of transcriptional activity across the entire embryo that becomes defined into discrete regions of high activity as cleavage continues (Jäckle et al. 1986). The critical element appears to be the expression of the Hunchback protein, which by the end of nuclear division cycle 12 is found at high levels across the anterior part of the embryo, and then forms a steep gradient through about 15 nuclei. The last third of the embryo has undetectable Hunchback levels. The transcription patterns of the anterior gap genes are initiated by the different concentrations of the Hunchback and Bicoid proteins. High levels of Hunchback protein induce the expression of giant, while the Krüppel transcript appears over the region where Hunchback begins to decline. High levels of Hunchback protein also prevent the transcription of the posterior gap genes (such as knirps) in the anterior part of the embryo (Struhl et al. 1992). It is thought that a gradient of the Caudal protein, highest at the posterior pole, is responsible for activating the abdominal gap genes knirps and giant. The giant gene has two methods for its activation, one for its anterior expression band and one for its posterior expression band (Rivera-Pomar 1995; Schulz and Tautz 1995).
After the establishment of these patterns by the maternal effect genes and Hunchback, the expression of each gap gene becomes stabilized and maintained by interactions between the different gap gene products themselves (Figure 9.21B).‡ For instance, Krüppel gene expression is negatively regulated on its anterior boundary by the Hunchback and Giant proteins and on its posterior boundary by the Knirps and Tailless proteins (Jäckle et al. 1986; Harding and Levine 1988; Hoch et al. 1992). If Hunchback activity is lacking, the domain of Krüppel expression extends anteriorly. If Knirps activity is lacking, Krüppel gene expression extends more posteriorly. The boundaries between the regions of gap gene transcription are probably created by mutual repression. Just as the Giant and Hunchback proteins can control the anterior boundary of Krüppel transcription, so Krüppel protein can determine the posterior boundaries of giant and hunchback transcription. If an embryo lacks the Krüppel gene, hunchback transcription continues into the area usually allotted to Krüppel (Jäckle et al. 1986; Kraut and Levine 1991). These boundary-forming inhibitions are thought to be directly mediated by the gap gene products, because all four major gap genes (hunchback, giant, Krüppel, and knirps) encode DNA-binding proteins that can activate or repress the transcription of other gap genes (Knipple et al. 1985; Gaul and Jäckle 1990; Capovilla et al. 1992).
The pair-rule genes
The first indication of segmentation in the fly embryo comes when the pair-rule genes are expressed during the thirteenth division cycle. The transcription patterns of these genes are striking in that they divide the embryo into the areas that are the precursors of the segmental body plan. As can be seen in Figure 9.22B-Eand Figure 9.8C, one vertical band of nuclei (the cells are just beginning to form) expresses a pair-rule gene, then another band of nuclei does not express it, and then another band of nuclei expresses it again. The result is a “zebra stripe” pattern along the anterior-posterior axis, dividing the embryo into 15 subunits (Hafen et al. 1984). Eight genes are currently known to be capable of dividing the early embryo in this fashion; they are listed in Table 9.2. It is important to note that not all nuclei express the same pair-rule genes. In fact, within each parasegment, each row of nuclei has its own constellation of pair-rule gene expression that distinguishes it from any other row.

Figure 9.22
Specific promoter regions of the even-skipped gene control specific transcription bands in the embryo. (A) Partial map of the eve promoter, showing the regions responsible for the various stripes. (B-E) A reporter β-galactosidase gene was fused (more...)
Table 9.2
Major genes affecting segmentation pattern in Drosophila.
How are some nuclei of the Drosophila embryo told to transcribe a particular gene while their neighbors are told not to transcribe it? The answer appears to come from the distribution of the protein products of the gap genes. Whereas the RNA of each of the gap genes has a very discrete distribution that defines abutting or slightly overlapping regions of expression, the protein products of these genes extend more broadly. In fact, they overlap by at least 8–10 nuclei (which at this stage accounts for about two to three segment primordia). This was demonstrated in a striking manner by Štanojevíc and co-workers (1989). They fixed cellularizing blastoderms (i.e., the stage when cells are beginning to form at the rim of the syncytial embryo), stained the Hunchback protein with an antibody carrying a red dye, and simultaneously stained the Krüppel protein with an antibody carrying a green dye. Cellularizing regions that contained both proteins bound both antibodies and were stained bright yellow (see Figure 9.8B). Krüppel protein overlaps with Knirps protein in a similar manner in the posterior region of the embryo (Pankratz et al. 1990).
Three genes are known to be the primary pair-rule genes. These genes—hairy, even-skipped, and runt—are essential for the formation of the periodic pattern, and they are directly controlled by the gap gene proteins. The enhancers of the primary pair-rule genes are recognized by gap gene proteins, and it is thought that the different concentrations of gap gene proteins determine whether a pair-rule gene is transcribed or not. The enhancers of the primary pair-rule genes are often modular: the control over each stripe is located in a discrete region of the DNA. One of the best-studied enhancers is that for the even-skipped gene. The structure of this enhancer is shown in Figure 9.22A. It is composed of modular units arranged in such a way that each unit regulates a separate stripe. For instance, the second even-skipped stripe is repressed by both Giant and Krüppel proteins and is activated by Hunchback protein and low concentrations of Bicoid (Figure 9.23; Small et al. 1991, 1992; Štanojevíc et al. 1991). DNase I footprinting (see Chapter 5) showed that the enhancer region for this stripe contains six binding sites for Krüppel protein, three for Hunchback protein, three for Giant protein, and five for Bicoid protein. Similarly, even-skipped stripe 5 is regulated negatively by Krüppel protein (on its anterior border) and by Giant protein (on its posterior border) (Small et al. 1996; Fujioka 1999).
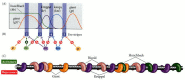
Figure 9.23
Hypothesis for the formation of the second stripe of transcription from the even-skipped gene. (A) The even-skipped gene is active where concentrations of most of the gap gene proteins are low. (B) Thus, the boundaries of eve transcription are determined (more...)
The importance of these enhancers can be shown by both genetic and biochemical means. First, a mutation in a particular enhancer can delete its particular stripe and no other. Second, if a reporter gene such as lacZ (encoding β-galactosidase) is fused to one of these enhancer elements, the lacZ gene is expressed only in that particular stripe (see Figure 9.22; Fujioka et al. 1999). Third, the placement of the stripes can be altered by deleting the gap genes that regulate them. Thus, the placement of the stripes of pair-rule gene expression is a result of (1) the modular cis-regulatory enhancer elements of the pair-rule genes and (2) the trans-regulatory gap gene proteins that bind to these enhancer sites.
Once initiated by the gap gene proteins, the transcription pattern of the primary pair-rule genes becomes stabilized by their interactions among themselves (Levine and Harding 1989). The primary pair-rule genes also form the context that allows or inhibits the expression of the later-acting secondary pair-rule genes. One such secondary pair-rule gene is fushi tarazu (ftz; Japanese, “too few segments;” Figures 9.8, 9.19, 9.20). Early in cycle 14, ftz RNA and protein are seen throughout the segmented portion of the embryo. However, as the proteins from the primary pair-rule genes begin to interact with the ftz enhancer, the ftz gene is repressed in certain bands of nuclei to create interstripe regions. Meanwhile, the Ftz protein interacts with its own promoter to stimulate more transcription of the ftz gene (Figure 9.24; Edgar et al. 1986b; Karr and Kornberg 1989; Schier and Gehring 1992).
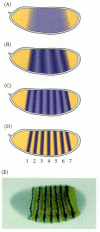
Figure 9.24
Transcription of the fushi tarazu gene in the Drosophila embryo. (A-D) At the beginning of cycle 14, there is low-level transcription of ftz in each of the nuclei in the segmented region of the embryo. Within the next 30 minutes, the expression pattern (more...)
The expression of the each pair-rule gene in seven stripes divides the embryo into fourteen parasegments, with each pair-rule gene being expressed in alternate parasegments. Moreover, each row of nuclei within each parasegment expresses a particular and unique combination of pair-rule products. These products will activate the next level of segmentation genes, the segment polarity genes.
The segment polarity genes
So far, our discussion has described interactions between molecules within the syncytial embryo. But once cells form, interactions take place between the cells. These intercellular interactions are mediated by the segment polarity genes, and they accomplish two important tasks. First, they reinforce the parasegmental periodicity established by the earlier transcription factors. Second, through this cell-to-cell signaling, cell fates are established within each parasegment.
The segment polarity genes encode proteins that are constituents of the Wingless and Hedgehog signal transduction pathways (see Chapter 6). Mutations in these genes lead to defects in segmentation and in gene expression pattern across each parasegment. The development of the normal pattern relies on the fact only one row of cells in each parasegment is permitted to express the Hedgehog protein, and only one row of cells in each parasegment is permitted to express the Wingless protein. The key to this pattern is the activation of the engrailed gene in those cells that are going to express the Hedgehog protein. The engrailed gene is activated when cells have high levels of the Even-skipped, Fushi tarazu, or Paired transcription factors. Moreover, it is repressed in those cells that receive high levels of Odd-skipped, Runt, or Sloppy-paired proteins. As a result, Engrailed is expressed in fourteen stripes across the anterior-posterior axis of the embryo (see Figure 9.8D). (Indeed, in mutations that cause the embryo to be deficient in Fushi tarazu, only seven bands of Engrailed are expressed.) These stripes of engrailed transcription mark the anterior boundary of each parasegment (and the posterior border of each segment). The wingless gene is activated in those bands of cells that receive little or no Even-skipped or Fushi tarazu proteins, but which do contain the Sloppy-paired protein. This causes wingless to be transcribed solely in the row of cells directly anterior to the cells where engrailed is transcribed (Figure 9.25).
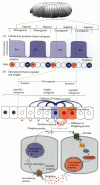
Figure 9.25
Model for the transcription of the segment polarity genes engrailed (en) and wingless (wg). (A) The expression of wg and en is initiated by pair-rule genes. The en gene is expressed when the cells contain high concentrations of either Even-skipped or (more...)
Once wingless and engrailed expression is established in adjacent cells, this pattern must be maintained to retain the parasegmental periodicity of the body plan established by the pair-rule genes. It should be remembered that the mRNAs and proteins involved in initiating these patterns are short-lived, and that the patterns must be maintained after their initiators are no longer being synthesized. The maintenance of these patterns is regulated by interactions between cells expressing wingless and those expressing engrailed. The Wingless protein, secreted from the wingless-expressing cells, diffuses to adjacent cells. The cells expressing engrailed can bind this protein because they contain the Drosophila membrane receptor protein for Wingless, D-Frizzled-2 (see Figure 6.23; Bhanot et al. 1996). This receptor activates the Wnt signal transduction pathway, resulting in the continued expression of engrailed (Siegfried et al. 1994).
Moreover, this activation starts another portion of this reciprocal pathway. The Engrailed protein activates the transcription of the hedgehog gene in the engrailed-expressing cells. The Hedgehog protein can bind to the Hedgehog receptor (the Patched protein) on neighboring cells. When it binds to the adjacent posterior cells, it stimulates the expression of the wingless gene. The result is a reciprocal loop wherein the Engrailed-synthesizing cells secrete the Hedgehog protein, which maintains the expression of the wingless gene in the neighboring cells, while the Wingless-secreting cells maintain the expression of the engrailed and hedgehog genes in their neighbors in turn (Heemskerk et al. 1991; Ingham et al. 1991; Mohler and Vani 1992). In this way, the transcription pattern of these two types of cells is stabilized. This interaction creates a stable boundary, as well as a signaling center from which Hedgehog and Wingless proteins diffuse across the parasegment.
The diffusion of these proteins is thought to provide the gradients by which the cells of the parasegment acquire their identities. This process can be seen in the dorsal epidermis, where the rows of larval cells produce different cuticular structures depending on their position within the segment. The 1° row consists of large, pigmented spikes called denticles. Posterior to these cells, the 2° row produces a smooth epidermal cuticle. The next two cell rows have a 3° fate, making small, thick hairs, and these are followed by several rows of cells that adopt the 4° fate, producing fine hairs (Figure 9.26).

Figure 9.26
Cell specification by the Wingless/Hedgehog signaling center. (A) Bright-field photograph of wild-type Drosophila embryo, showing the position of the third abdominal segment. (B) Close-up of the dorsal area of the A3 segment, showing the different cuticular (more...)
The wingless-expressing cells lie within the region producing the fine hairs, while the hedgehog-expressing cells are near the 1° row of cells. The fates of the cells can be altered by experimentally increasing or decreasing the levels of Hedgehog or Wingless protein (Heemskerk and DiNardo 1994;Bokor and DiNardo 1996; Porter et al. 1996). For example, if the hedgehog gene is fused to a heat shock promoter and the embryos are grown at a temperature that activates the gene, more Hedgehog protein is made, and the cells normally showing 3° fates will become 2° cells. The rows of 4° cells farthest from the Wingless-secreting cells may also become 3° or 2° cells. It seems that the cells closest to the Wingless secreters cannot respond to Hedgehog, and Hedgehog cannot, by itself, specify the 1° fate (which may require the expression of certain pair-rule gene products). Thus, Hedgehog and Wingless appear necessary for elaborating the entire pattern of cell types across the parasegment. However, the mechanism by which they accomplish this specification is not clear. Either these signals act in a graded fashion, as morphogens, or they act locally to initiate a cascade of local signaling events, in which each interaction uses a different ligand and receptor (Figure 9.26). The resulting pattern of cell fates also changes the focus of patterning from parasegment to segment. There are now external markers, as the engrailed-expressing cells become the most posterior cells of each segment.
WEBSITE
9.7 Asymmetrical spread of morphogens. It is unlikely that morphogens such as Wingless spread by free diffusion. The asymmetry of Wingless diffusion suggests that neighboring cells play a crucial role in moving the protein. http://www.devbio.com/chap09/link0907.shtml
WEBSITE
9.8 Getting a head in the fly. The segment polarity genes may act differently in the head than in the trunk. Indeed, the formation of the Drosophila head may differ significantly from the way the rest of the body is formed. http://www.devbio.com/chap09/link0908.shtml
The Homeotic Selector Genes
Patterns of homeotic gene expression
After the segmental boundaries have been established, the characteristic structures of each segment are specified. This specification is accomplished by the homeotic selector genes (Lewis 1978). There are two regions of Drosophila chromosome 3 that contain most of these homeotic genes (Figure 9.27). One region, the Antennapedia complex, contains the homeotic genes labial (lab), Antennapedia (Antp), sex combs reduced (scr), deformed (dfd), and proboscipedia (pb). The labial and deformed genes specify the head segments, while sex combs reduced and Antennapedia contribute to giving the thoracic segments their identities. The proboscipedia gene appears to act only in adults, but in its absence, the labial palps of the mouth are transformed into legs (Wakimoto et al. 1984; Kaufman et al. 1990). The second region of homeotic genes is the bithorax complex (Lewis 1978). There are three protein-coding genes found in this complex: ultrabithorax (ubx), which is required for the identity of the third thoracic segment; and the abdominal A (abdA) and Abdominal B (AbdB) genes, which are responsible for the segmental identities of the abdominal segments (Sánchez-Herrero et al. 1985). The lethal phenotype of the triple-point mutant Ubx-, abdA-, AbdB- is identical to that resulting from a deletion of the entire bithorax complex (Casanova et al. 1987). The chromosome region containing both the Antennapedia complex and the bithorax complex is often referred to as the homeotic complex (Hom-C).
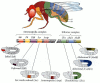
Figure 9.27
Homeotic gene expression in Drosophila. In the center are the genes of the Antennapedia and bithorax complexes and their functional domains. Below and above the gene map are the regions of homeotic gene expression (both mRNA and protein) in the blastoderm (more...)
Because these genes are responsible for the specification of fly body parts, mutations in them lead to bizarre phenotypes. In 1894, William Bateson called these organisms “homeotic mutants,” and they have fascinated developmental biologists for decades.§ For example, the body of the normal adult fly contains three thoracic segments, all of which produce a pair of legs. The first thoracic segment does not produce any further appendages, but the second thoracic segment produces both a set of legs and a set of wings. The third thoracic segment produces a set of wings and a set of balancers known as halteres. In homeotic mutants, these specific segmental identities can be changed. When the ultrabithorax gene is deleted, the third thoracic segment (which is characterized by halteres) becomes transformed into another second thoracic segment. The result (Figure 9.28) is a fly with four wings—an embarrassing situation for a classic dipteran. ¶ Similarly, the Antennapedia protein is usually used to specify the second thoracic segment of the fly. But when flies have a mutation wherein the Antennapedia gene is expressed in the head (as well as in the thorax), legs rather than antennae grow out of the head sockets (Figure 9.29). In the recessive mutant of Antennapedia, the gene fails to be expressed in the second thoracic segment, and antennae sprout out of the leg positions (Struhl 1981; Frischer et al. 1986; Schneuwly et al. 1987).

Figure 9.28
This four-winged fruit fly was constructed by putting together three mutations in cis regulators of the ultrabithorax gene. These mutations effectively transform the third thoracic segment into another second thoracic segment (i.e., halteres into wings). (more...)

Figure 9.29
(A) Head of a wild-type fly. (B) Head of a fly containing the Antennapedia mutation that converts antennae into legs. (From Kaufman et al. 1990; photographs courtesy of T. C. Kaufman.)
These major homeotic selector genes have been cloned and their expression analyzed by in situ hybridization (Harding et al. 1985; Akam 1987). Transcripts from each gene can be detected in specific regions of the embryo and are especially prominent in the central nervous system (see Figure 9.27).
Initiating the patterns of homeotic gene expression
The initial domains of homeotic gene expression are influenced by the gap genes and pair-rule genes. For instance, the expression of the abdA and AbdB genes is repressed by the gap gene proteins Hunchback and Krüppel. This inhibition prevents these abdomen-specifying genes from being expressed in the head and thorax (Casares and Sánchez-Herrero 1995). Conversely, the Ultrabithorax gene is activated by certain levels of the Hunchback protein, so that it is originally transcribed in a broad band in the middle of the embryo, and the transcription of Antennapedia is activated by Krüppel (Harding and Levine 1988; Struhl et al. 1992). The boundaries of homeotic gene expression are soon confined to the parasegments defined by the Fushi tarazu and Even-skipped proteins (Ingham and Martinez-Arias 1986; Müller and Bienz 1992).
Maintaining the patterns of homeotic gene expression
The expression of homeotic genes is a dynamic process. The Antennapedia gene (Antp), for instance, although initially expressed in presumptive parasegment 4, soon appears in parasegment 5. As the germ band expands, Antp expression is seen in the presumptive neural tube as far posterior as parasegment 12. During further development, the domain of Antp expression contracts again, and Antp transcripts are localized strongly to parasegments 4 and 5. Like that of other homeotic genes, Antp expression is negatively regulated by all the homeotic gene products expressed posterior to it (Harding and Levine 1989; González-Reyes and Morata 1990). In other words, each of the bithorax complex genes represses the expression of Antennapedia. If the Ultrabithorax gene is deleted, Antp activity extends through the region that would normally have expressed Ubx and stops where the Abd region begins. (This allows the third thoracic segment to form wings like the second thoracic segment, as in Figure 9.29.) If the entire bithorax complex is deleted, Antp expression extends throughout the abdomen. (Such a larva does not survive, but the cuticle pattern throughout the abdomen is that of the second thoracic segment.)
As we saw above, the gap gene and pair-rule gene proteins are transient, but the identities of the segments must be stabilized so that differentiation can occur. Thus, once the transcription patterns of the homeotic genes have become stabilized, they are “locked” into place by alteration of the chromatin conformation in these genes. The repression of homeotic genes appears to be maintained by the Polycomb family of proteins, while the active chromatin conformation appears to be maintained by the Trithorax proteins (Ingham and Whittle 1980; McKeon and Brock 1991; Simon et al. 1992).
Realisator genes
The search is now on for “realisator genes,” those genes that are the targets of the homeotic gene proteins and which function to form the specified tissue or organ primordia. In the formation of the second thoracic segment, for example, Antennapedia is expressed. Casares and Mann (1998) have shown that Antennapedia protein binds to the enhancer of the homothorax gene and prevent its expression. Homothorax is necessary for producing a transcription factor critical for antenna formation. Therefore, one of Antennapedia's functions is to suppress those genes necessary for antenna development.
The Ultrabithorax protein is able to repress the expression of the Wingless gene in those cells that will become the halteres of the fly. One of the major differences between the appendage-forming cells of the second and the third thoracic segments is that Wingless expression occurs in the appendage-forming cells of the second thoracic segment, but not in those of the third thoracic segment. Wingless acts as a growth promoter and morphogen in these tissues. In the third thoracic segment, Ubx protein is found in these cells, and it prevents the expression of the Wingless gene (Figure 9.30; Weatherbee et al. 1998). Thus, one of the ways in which Ubx protein specifies the third thoracic segment is by preventing the expression of those genes that would generate the wing tissue.

Figure 9.30
Antibody staining of the Ultrabithorax protein in (A) the wing disc and (B) the haltere disc of third instar Drosophila larvae. These are the cells that will give rise to a wing and a haltere, respectively. In the wing disc, Ultrabithorax staining can (more...)
Another target of the homeotic proteins, the distal-less gene (itself a homeobox-containing gene: see Sidelights and Speculations), is necessary for limb development and is active solely in the thorax. Distal-less expression is repressed in the abdomen, probably by a combination of Ubx and AbdA proteins that can bind to its enhancer and block transcription (Vachon et al. 1992; Castelli-Gair and Akam 1995). This presents a paradox, since parasegment 5 (entirely thoracic and leg-producing) and parasegment 6 (which includes most of the legless first abdominal segment) both expressUbx. How can these two very different segments be specified by the same gene? Castelli-Gair and Akam (1995) have shown that the mere presence of Ubx protein in a group of cells is not sufficient for specification. Rather, the time and place of its expression within the parasegment can be critical. Before Ubx expression, parasegments 4–6 have similar potentials. At division cycle 10, Ubx expression in the anterior parts of parasegments 5 and 6 prevents those parasegments from forming structures (such as the anterior spiracle) characteristic of parasegment 4. Moreover, in the posterior compartment of parasegment 6 (but not parasegment 5), Ubx protein blocks the formation of the limb primordium by repressing the distal-less gene. At division cycle 11, by which time Ubx has pervaded all of parasegment 6, the distal-less gene has become self-regulatory and cannot be repressed by Ubx (Figure 9.31).
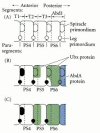
Figure 9.31
Schematic representation of the differences between Ubx expression in parasegments 5 and 6. (A) Before Ubx expression, each parasegment is competent to make both spiracles and legs. (B) At division cycle 10, early Ubx expression blocks the formation of (more...)
The Homeodomain Proteins
Homeodomain proteins are a family of transcription factors characterized by a 60-amino acid domain (the homeodomain) that binds to certain regions of DNA. The homeodomain was first discovered in those proteins whose absence or misregulation caused homeotic transformations of Drosophila segments. It is thought that homeodomain proteins activate batteries of genes that specify the particular properties of each segment. The homeodomain proteins include the products of the eight genes of the homeotic complex, as well as other proteins such as Fushi tarazu, Caudal, Distal-less and Bicoid. Homeodomain proteins are important in determining the anterior-posterior axes of both invertebrates and vertebrates. In Drosophila, the presence of certain homeodomain proteins is also necessary for the determination of specific neurons. Without these transcription factors, the fates of these neuronal cells are altered (Doe et al. 1988).
The homeodomain is encoded by a 180-base-pair DNA sequence known as the homeobox. The homeodomains appear to be the sites of these proteins that bind DNA, and they are critical in specifying cell fates. For instance, if a chimeric protein is constructed mostly of Antennapedia but with the carboxyl terminus (including the homeodomain) of Ultrabithorax, it can substitute for Ultrabithorax and specify the appropriate cells as parasegment 6 (Mann and Hogness 1990). The isolated homeodomain of Antennapedia will bind to the same promoters as the entire Antennapedia protein, indicating that the binding of this protein is dependent on its homeodomain (Müller et al. 1988).
The homeodomain folds into three α helices, the latter two folding into a helix-turn-helix conformation that is characteristic of transcription factors that bind DNA in the major groove of the double helix (Otting et al. 1990; Percival-Smith et al. 1990). The third helix is the recognition helix, and it is here that the amino acids make contact with the bases of the DNA. A four-base motif, TAAT, is conserved in nearly all sites recognized by homeodomains; it probably distinguishes those sites to which homeodomain proteins can bind. The 5´ terminal T appears to be critical in this recognition, as mutating it destroys all homeodomain binding. The base pairs following the TAAT motif are important in distinguishing between similar recognition sites. For instance, the next base pair is recognized by amino acid 9 of the recognition helix. Mutation studies have shown that the Bicoid and Antennapedia homeodomain proteins use lysine and glutamine, respectively, at position 9 to distinguish related recognition sites. The lysine of the Bicoid homeodomain recognizes the G of CG pairs, while the glutamine of the Antennapedia homeodomain recognizes the A of AT pairs (Figure 9.32; Hanes and Brent 1991). If the lysine in Bicoid is replaced by glutamine, the resulting protein will recognize Antennapedia-binding sites (Hanes and Brent 1989, 1991). Other homeodomain proteins show a similar pattern, in which one portion of the homeodomain recognizes the common TAAT sequence, while another portion recognizes a specific structure adjacent to it.
Cofactors for the Hom-C Genes
The genes of the Drosophila homeotic complex specify segmental fates, but they may need some help in doing it. The DNA-binding sites recognized by the homeodomains of the Hom-C proteins are very similar, and there is some overlap in their binding specificity. In 1990, Peifer and Wieschaus discovered that the product of the Extradenticle (Exd) gene interacts with several Hom-C proteins and may help specify the segmental identities. For instance, the Ubx protein is responsible for specifying the identity of the first abdominal segment (A1). Without Extradenticle protein, it will transform this segment into A3. Moreover, the Exd and Ubx proteins are both needed for the regulation of the decapentaplegic gene, and the structure of the decapentaplegic promoter suggests that the Extradenticle protein may dimerize with the Ubx protein on the enhancer of this target gene (Raskolb and Wieschaus 1994; van Dyke and Murre 1994). The Extradenticle protein includes a homeodomain, and the human protein PBX1, which resembles the Extradenticle protein, may play a similar role as a cofactor for human homeotic genes.
The product of the teashirt gene may also be an important cofactor. This zinc finger transcription factor is necessary for the functioning of the Sex combs reduced protein, which distinguishes between the labial and first thoracic segments. It is critical for the specification of the anterior prothoracic (parasegment 3) identity, and it may be the gene that specifies the “groundstate condition” of the homeotic complex. If the bithorax complex and the Antennapedia gene are removed, all the segments become anterior prothorax. The product of the teashirt gene appears to work with the Scr protein to distinguish thorax from head and to work throughout the trunk to prevent head structures from forming (Roder et al. 1992). ▪
WEBSITE
9.9 Homeotic genes and their protein products. The Hom-C genes have fascinating structures. The protein products of these genes bind to DNA in the presence of other proteins that may allow them to recognize specific sequences of DNA. http://www.devbio.com/chap09/link0909.shtml

Figure 9.32
Homeodomain-DNA interactions. (A) Homeodomain helix-turn-helix sequence within the major groove of the DNA. (B) Proposed pairing between the lysine of the Bicoid homeodomain and the CG base pair of its recognition sequence, and between the glutamine of (more...)
Footnotes
- *
Like the placement of the bicoid message, the location of the nanos message is determined by its 3´ untranslated region. If the bicoid 3´ UTR is experimentally placed on the protein-encoding region of nanos RNA, the nanos message gets placed in the anterior of the egg. When the RNA is translated, the Nanos protein inhibits the translation of hunchback and bicoid mRNAs, and the embryo forms two abdomens—one in the anterior of the embryo and one in the posterior (Gavis and Lehmann 1992). The localization of nanos RNA is ultimately dependent on interactions between the oocyte and the neighboring follicle cells that localize the oskar message to the posterior pole.
- †
Aficionados of information theory will recognize that the process by which the anterior-posterior information in morphogenetic gradients is transferred to discrete and different parasegments represents a transition from analog to digital specification. Specification is analog, determination digital. This process enables the transient information of the gradients in the syncytial blastoderm to be stabilized so that it can be utilized much later in development (Baumgartner and Noll 1990).
- ‡
The interactions between genes and gene products are facilitated by the fact that these reactions occur within a syncytium, in which the cell membranes have not yet formed.
- §
Homeo means “similar.” Homeotic mutants are mutants in which one structure is replaced by another (as where an antenna is replaced by a leg). Homeotic genes are genes whose mutation can cause such transformations; thus, they are genes that specify the identity of a particular body segment. The homeobox is a conserved DNA sequence of about 180 base pairs that is shared by many homeotic genes. This sequence encodes the 60-amino acid homeodomain, which recognizes specific DNA sequences. The homeodomain is an important region of the transcription factors encoded by homeotic genes (see Sidelights & Speculations). Not all genes with homeoboxes are homeotic genes, however.
- ¶
Dipterans (two-winged insects such as flies) are thought to have evolved from four-winged insects; it is possible that this change arose via alterations in the bithorax complex. Chapter 22 includes more speculation on the relationship between the homeotic complex and evolution.
- The Origins of Anterior-Posterior Polarity - Developmental BiologyThe Origins of Anterior-Posterior Polarity - Developmental Biology
Your browsing activity is empty.
Activity recording is turned off.
See more...